Summary information and primary citation
- PDB-id
- 7v59; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- RNA binding protein-RNA-DNA
- Method
- cryo-EM (5.26 Å)
- Summary
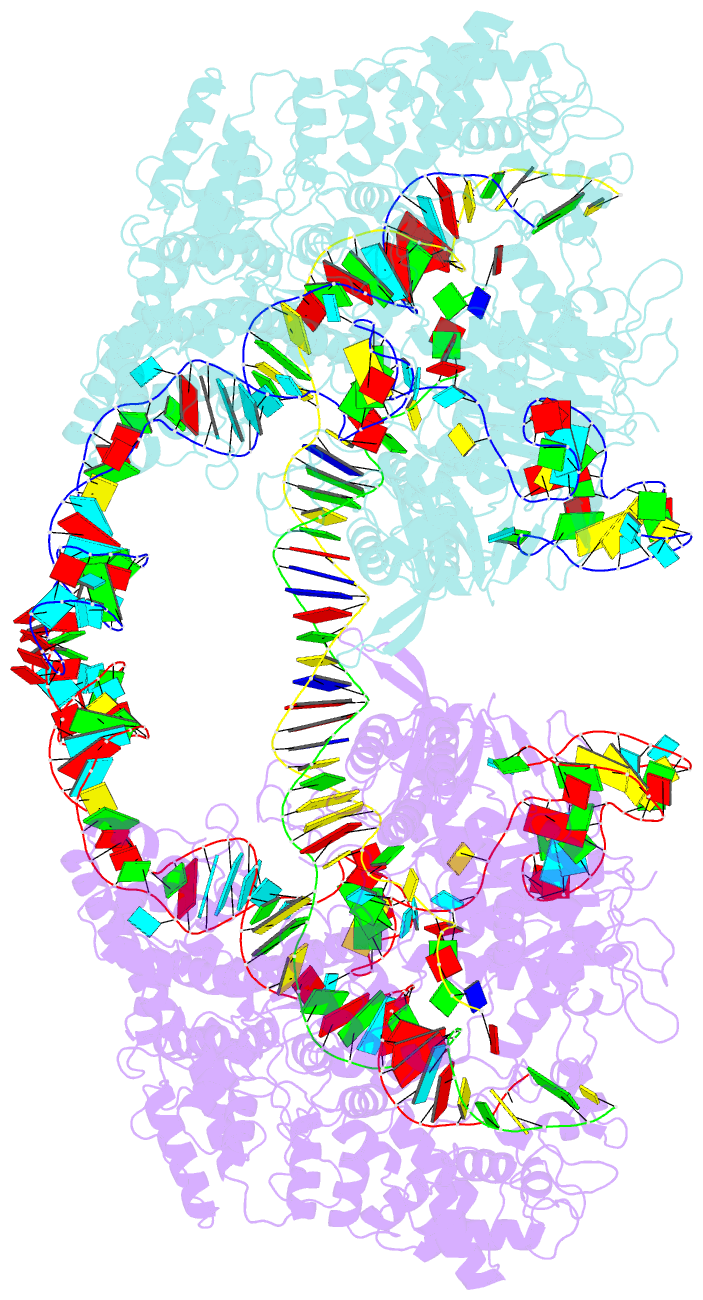
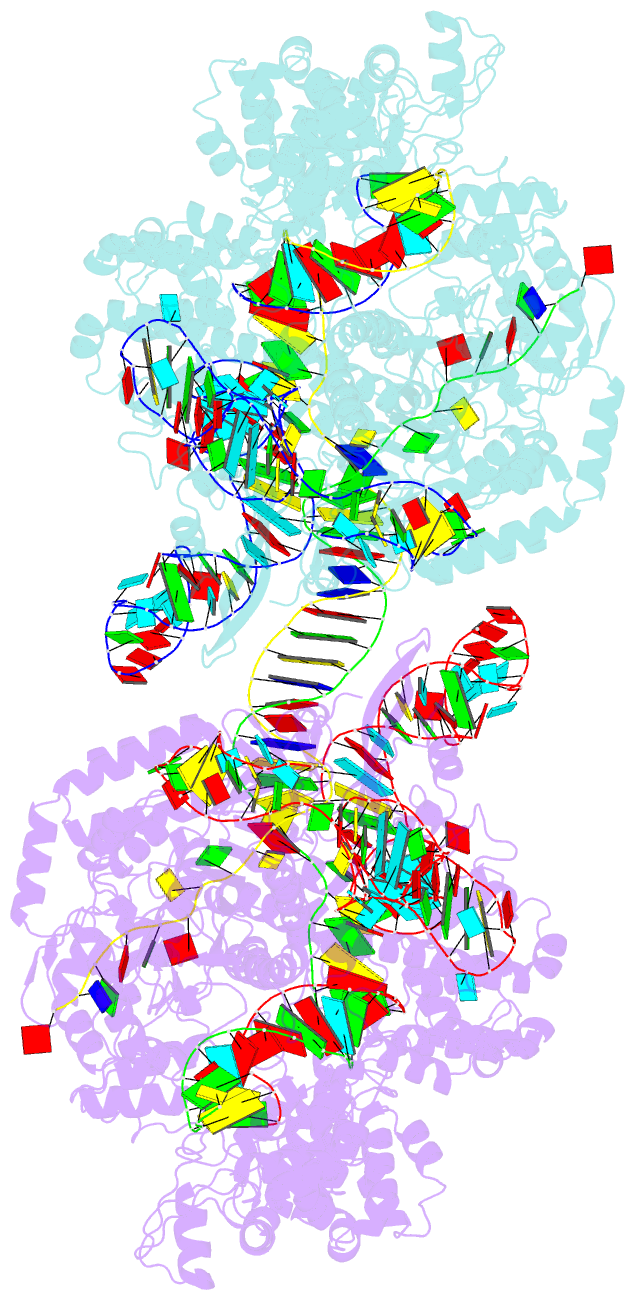
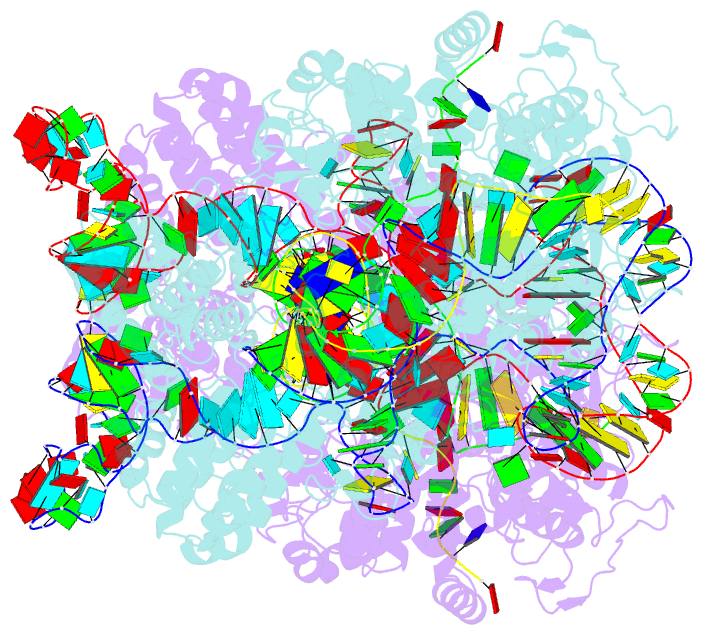
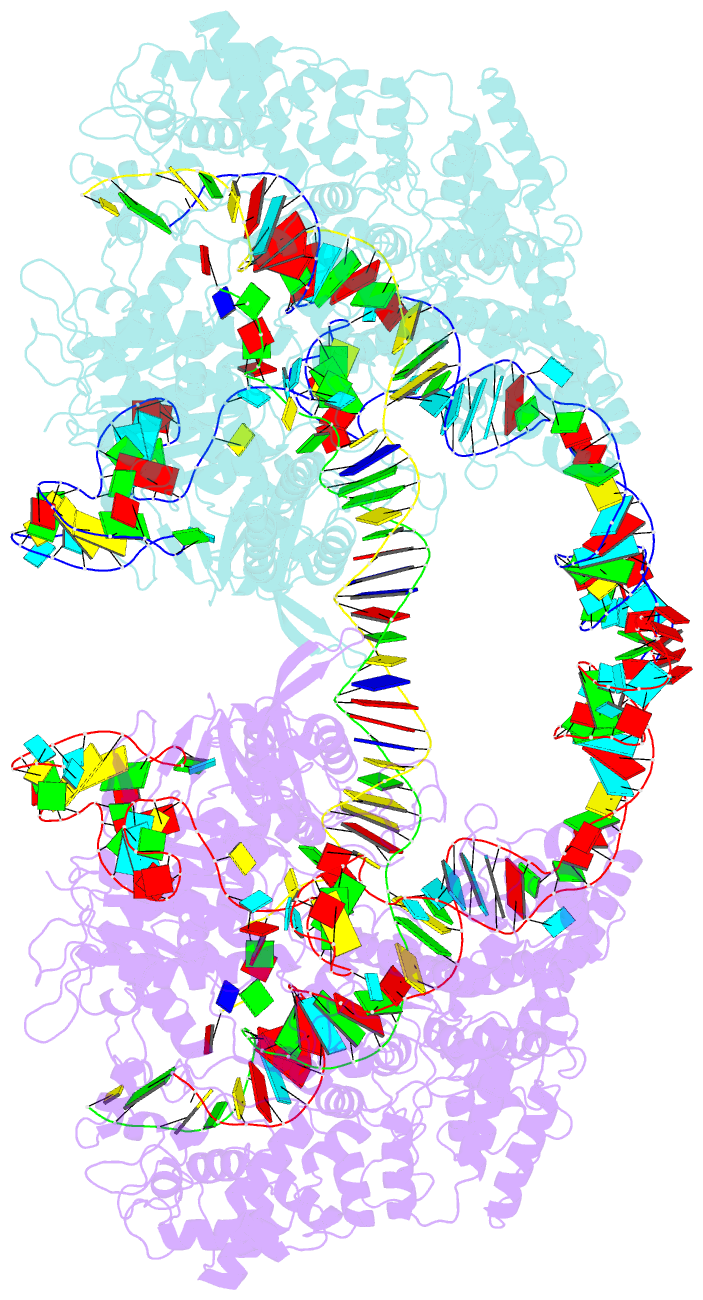
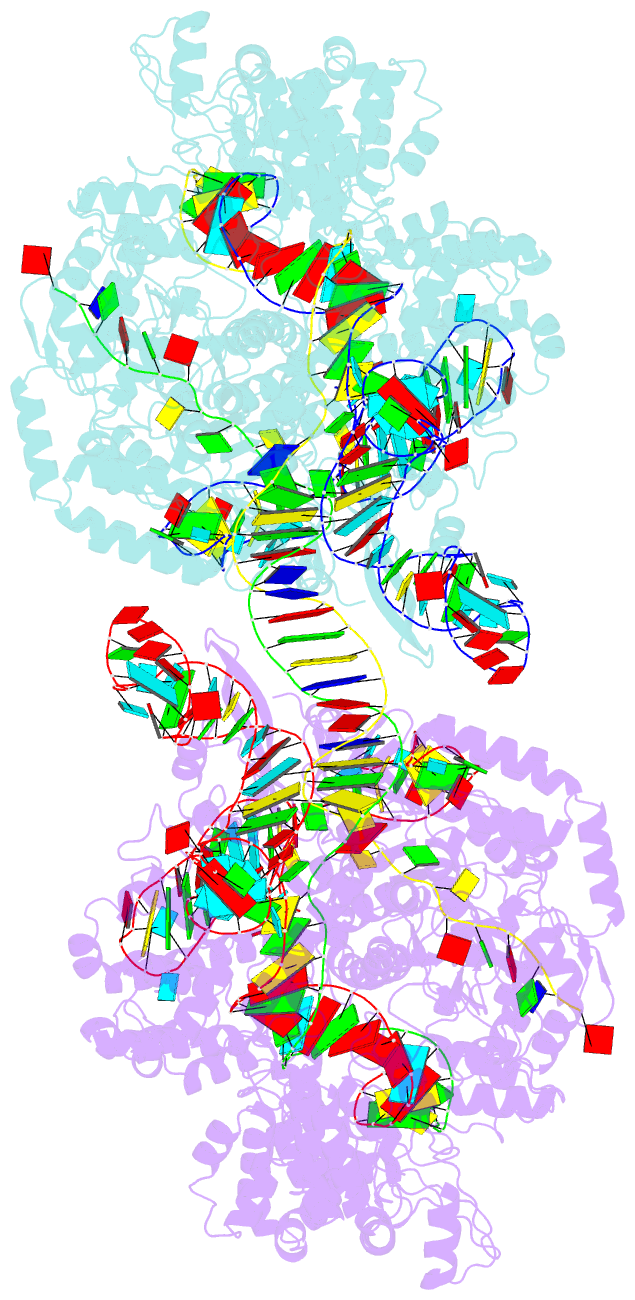
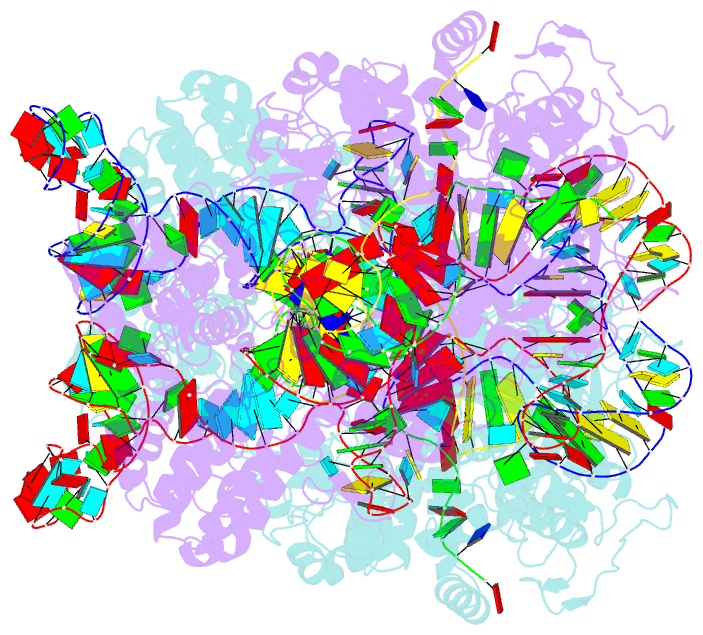
- cryo-EM structure of spycas9-sgrna-DNA dimer
- Reference
- Yang M, Sun R, Deng P, Yang Y, Wang W, Liu JG, Chen C (2021): "Nonspecific interactions between SpCas9 and dsDNA sites located downstream of the PAM mediate facilitated diffusion to accelerate target search." Chem Sci, 12, 12776-12784. doi: 10.1039/d1sc02633j.
- Abstract
- RNA-guided Streptococcus pyogenes Cas9 (SpCas9) is a sequence-specific DNA endonuclease that works as one of the most powerful genetic editing tools. However, how Cas9 locates its target among huge amounts of dsDNAs remains elusive. Here, combining biochemical and single-molecule fluorescence assays, we revealed that Cas9 uses both three-dimensional and one-dimensional diffusion to find its target with high efficiency. We further observed surprising apparent asymmetric target search regions flanking PAM sites on dsDNA under physiological salt conditions, which accelerates the target search efficiency of Cas9 by ∼10-fold. Illustrated by a cryo-EM structure of the Cas9/sgRNA/dsDNA dimer, non-specific interactions between DNA ∼8 bp downstream of the PAM site and lysines within residues 1151-1156 of Cas9, especially lys1153, are the key elements to mediate the one-dimensional diffusion of Cas9 and cause asymmetric target search regions flanking the PAM. Disrupting these non-specific interactions, such as mutating these lysines to alanines, diminishes the contribution of one-dimensional diffusion and reduces the target search rate by several times. In addition, low ionic concentrations or mutations on PAM recognition residues that modulate interactions between Cas9 and dsDNA alter apparent asymmetric target search behaviors. Together, our results reveal a unique searching mechanism of Cas9 under physiological salt conditions, and provide important guidance for both in vitro and in vivo applications of Cas9.