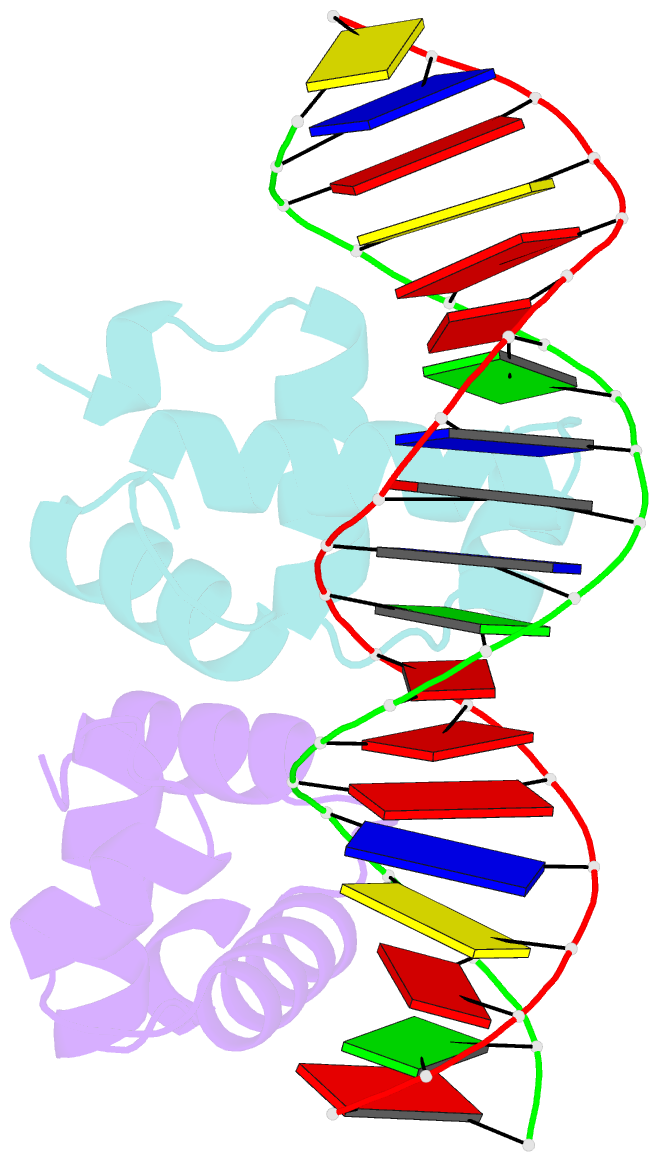
Summary information and primary citation
- PDB-id
- 7ve5; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- DNA binding protein-DNA
- Method
- X-ray (2.0 Å)
- Summary
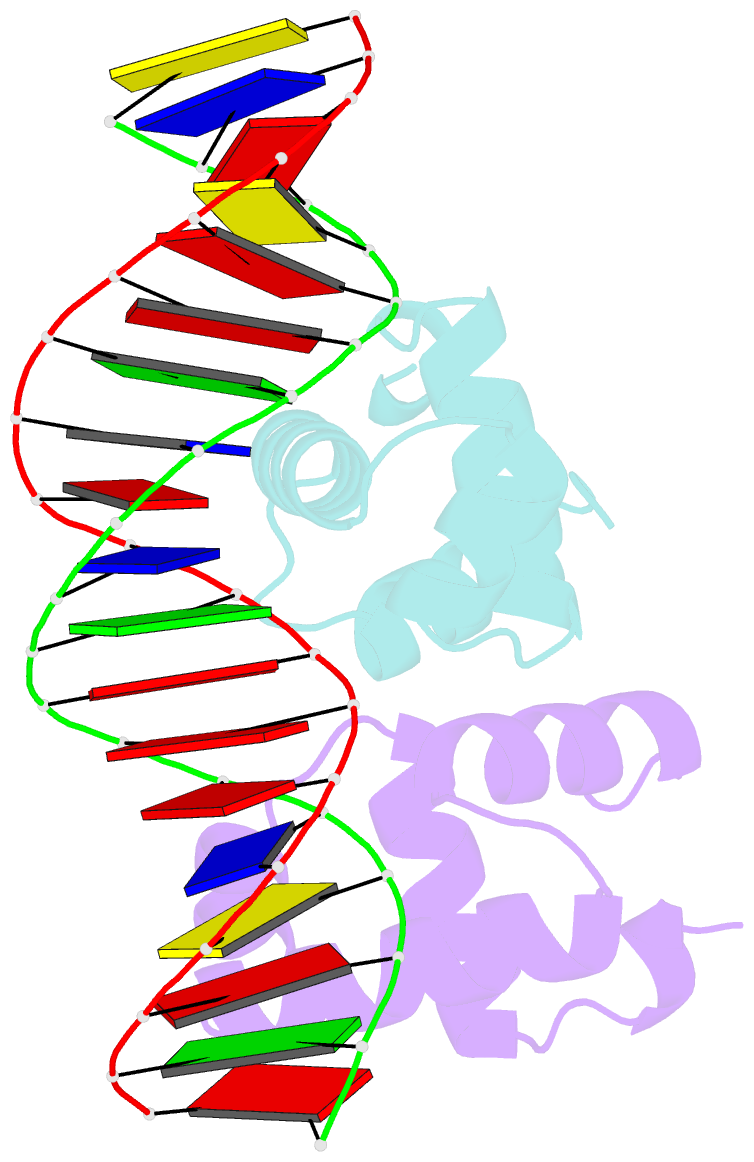
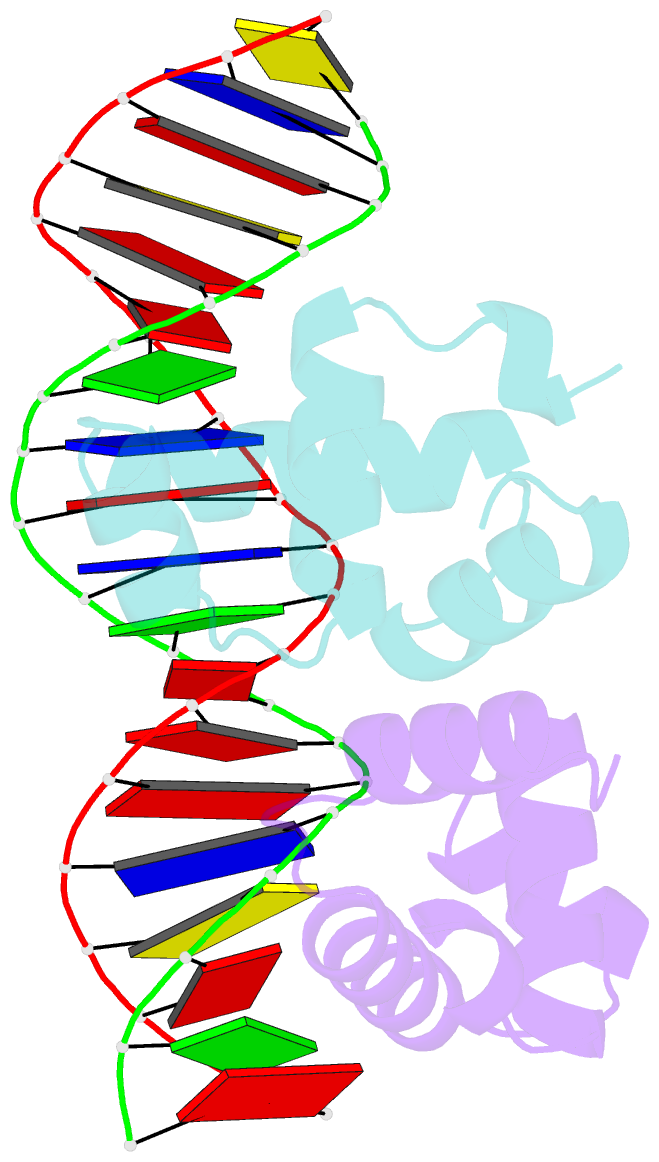
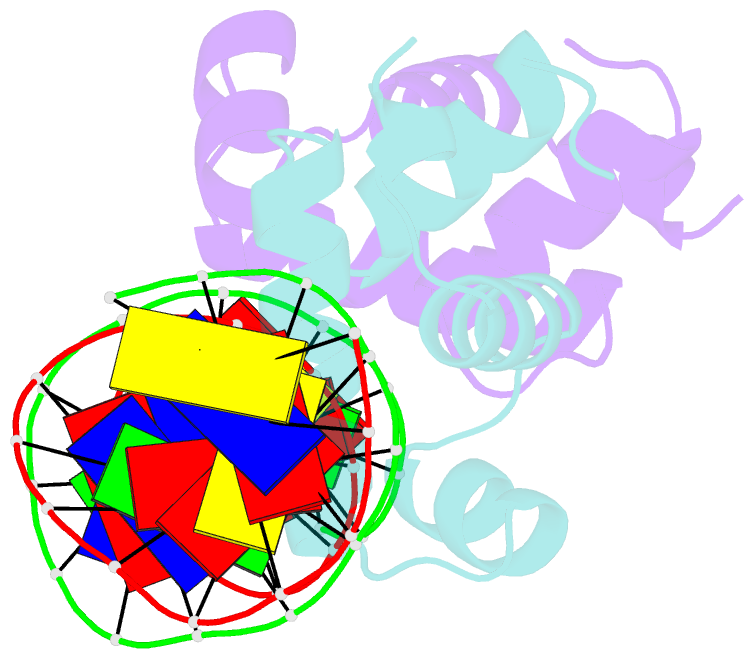
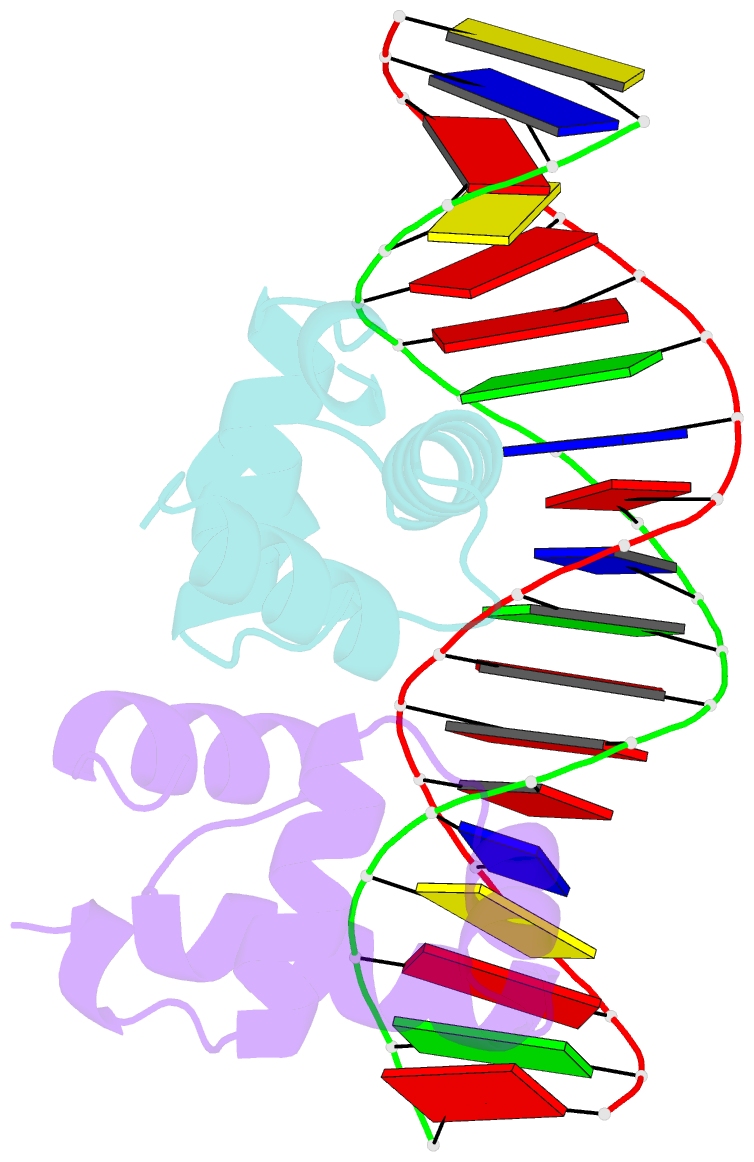
- C-terminal domain of vrar
- Reference
- Kumar JV, Tseng TS, Lou YC, Wei SY, Wu TH, Tang HC, Chiu YC, Hsu CH, Chen C (2022): "Structural insights into DNA binding domain of vancomycin-resistance-associated response regulator in complex with its promoter DNA from Staphylococcus aureus." Protein Sci., 31, e4286. doi: 10.1002/pro.4286.
- Abstract
- In Staphylococcus aureus, vancomycin-resistance-associated response regulator (VraR) is a part of the VraSR two-component system, which is responsible for activating a cell wall-stress stimulon in response to an antibiotic that inhibits cell wall formation. Two VraR-binding sites have been identified: R1 and R2 in the vraSR operon control region. However, the binding of VraR to a promoter DNA enhancing downstream gene expression remains unclear. VraR contains a conserved N-terminal receiver domain (VraRN ) connected to a C-terminal DNA binding domain (VraRC ) with a flexible linker. Here, we present the crystal structure of VraRC alone and in complex with R1-DNA in 1.87- and 2.0-Å resolution, respectively. VraRC consisting of four α-helices forms a dimer when interacting with R1-DNA. In the VraRC -DNA complex structure, Mg2+ ion is bound to Asp194. Biolayer interferometry experiments revealed that the addition of Mg2+ to VraRC enhanced its DNA binding affinity by eightfold. In addition, interpretation of NMR titrations between VraRC with R1- and R2-DNA revealed the essential residues that might play a crucial role in interacting with DNA of the vraSR operon. The structural information could help in designing and screening potential therapeutics/inhibitors to deal with antibiotic-resistant S. aureus via targeting VraR.