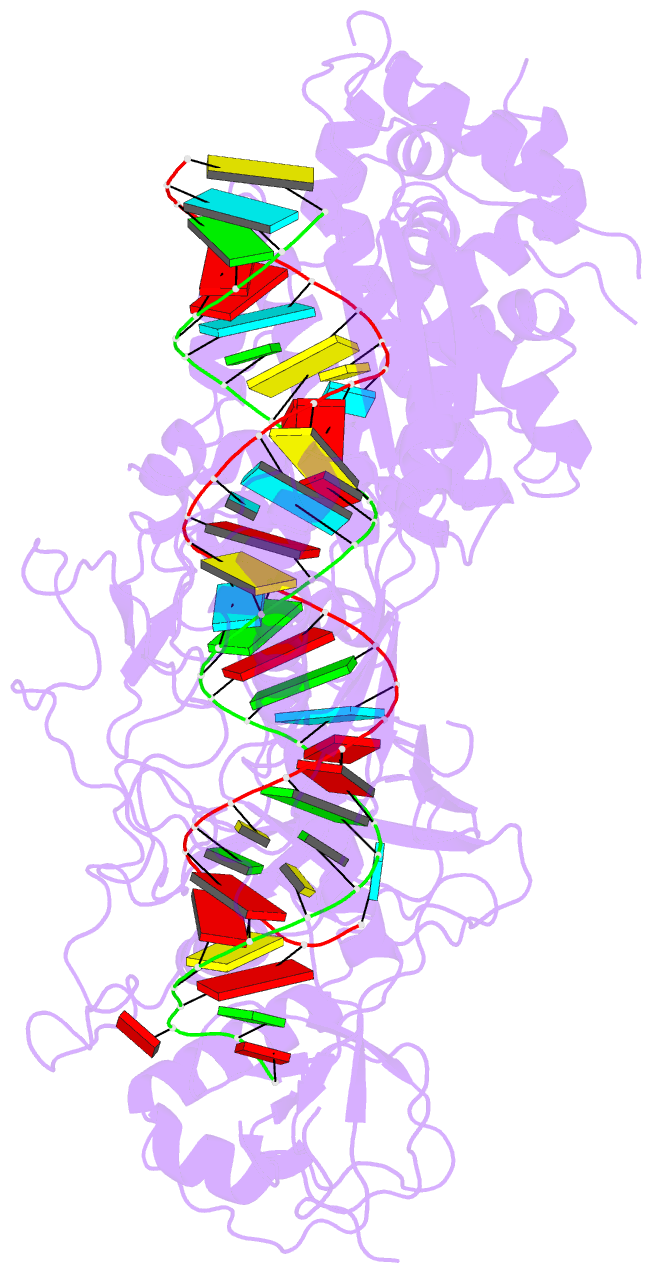
Summary information and primary citation
- PDB-id
- 7vg3; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- RNA binding protein-RNA
- Method
- cryo-EM (3.73 Å)
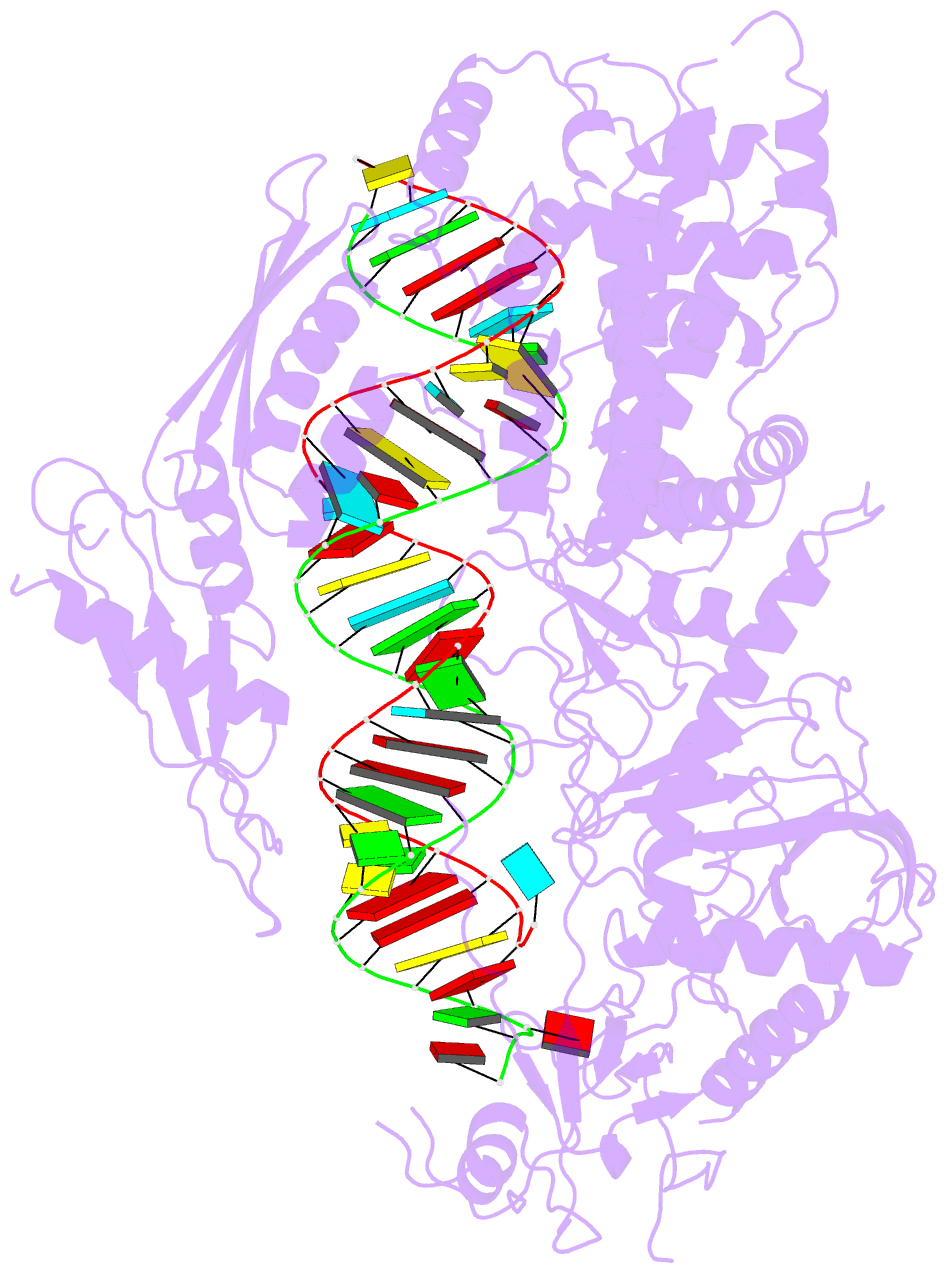
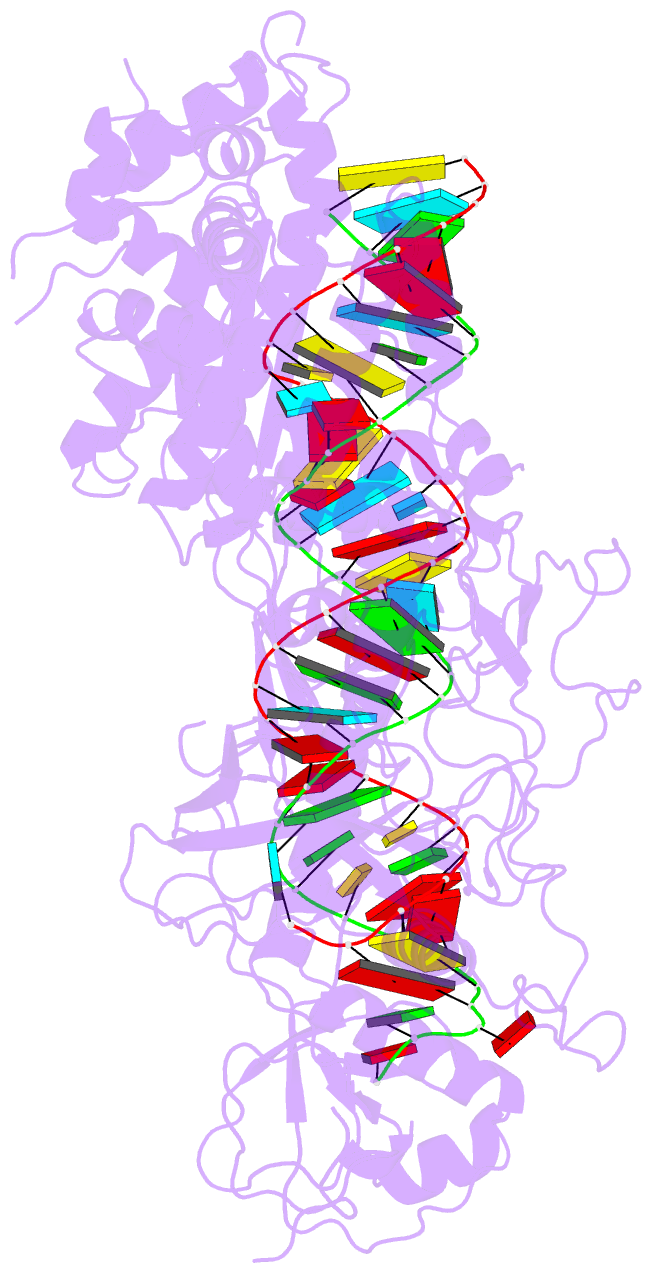
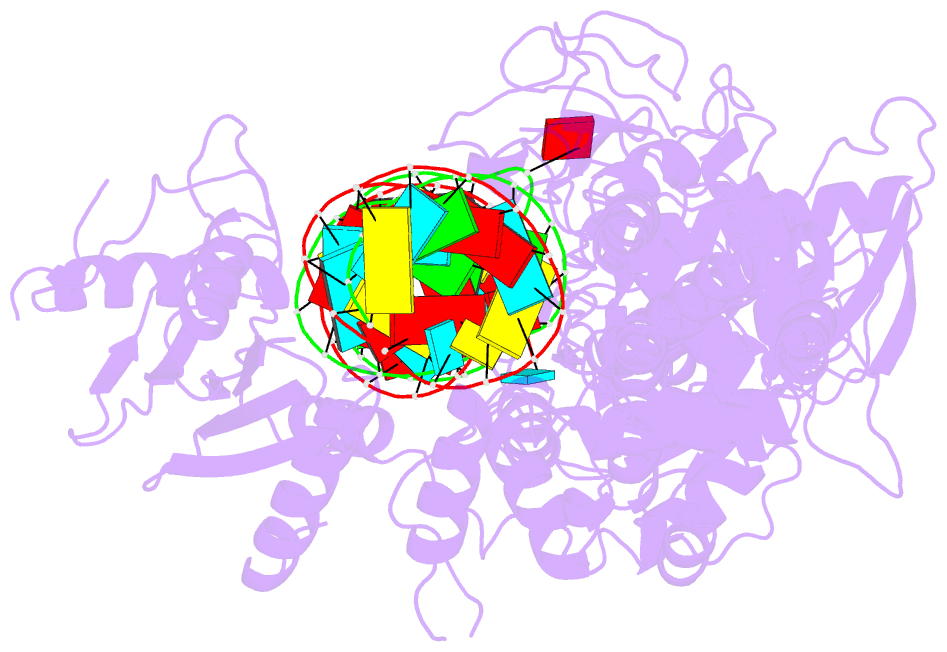
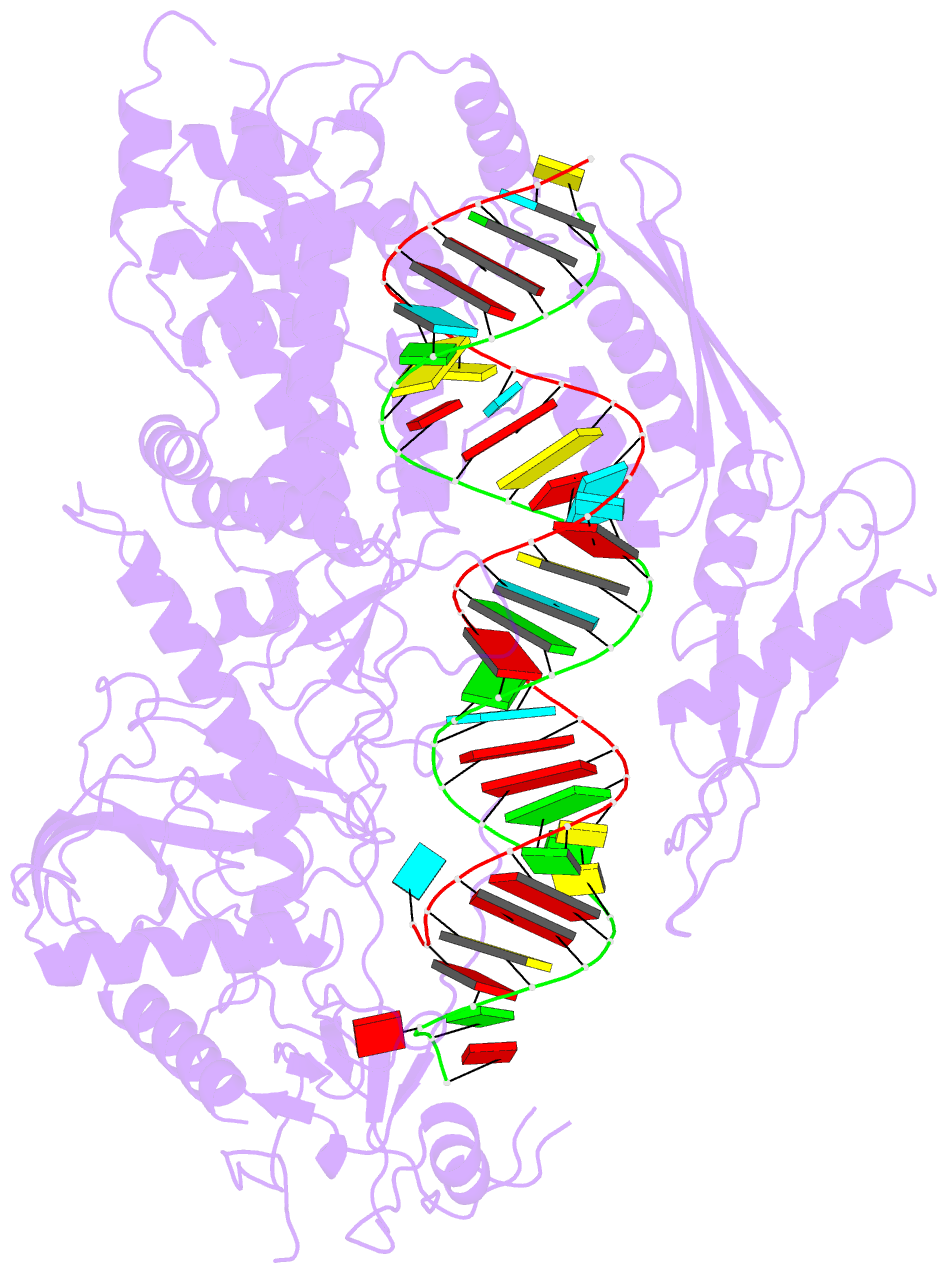
- Summary
- cryo-EM structure of arabidopsis dcl3 in complex with a 30-bp RNA
- Reference
- Wang Q, Xue Y, Zhang L, Zhong Z, Feng S, Wang C, Xiao L, Yang Z, Harris CJ, Wu Z, Zhai J, Yang M, Li S, Jacobsen SE, Du J (2021): "Mechanism of siRNA production by a plant Dicer-RNA complex in dicing-competent conformation." Science, 374, 1152-1157. doi: 10.1126/science.abl4546.
- Abstract
- In eukaryotes, small RNAs (sRNAs) play critical roles in multiple biological processes. Dicer endonucleases are a central part of sRNA biogenesis. In plants, DICER-LIKE PROTEIN 3 (DCL3) produces 24-nucleotide (nt) small interfering RNAs (siRNAs) that determine the specificity of the RNA-directed DNA methylation pathway. Here, we determined the structure of a DCL3–pre-siRNA complex in an active dicing-competent state. The 5′-phosphorylated A1 of the guide strand and the 1-nt 3′ overhang of the complementary strand are specifically recognized by a positively charged pocket and an aromatic cap, respectively. The 24-nt siRNA length dependence relies on the separation between the 5′-phosphorylated end of the guide RNA and dual cleavage sites formed by the paired ribonuclease III domains. These structural studies, complemented by functional data, provide insight into the dicing principle for Dicers in general.