Summary information and primary citation
- PDB-id
- 7vjm; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription-DNA
- Method
- X-ray (3.0 Å)
- Summary
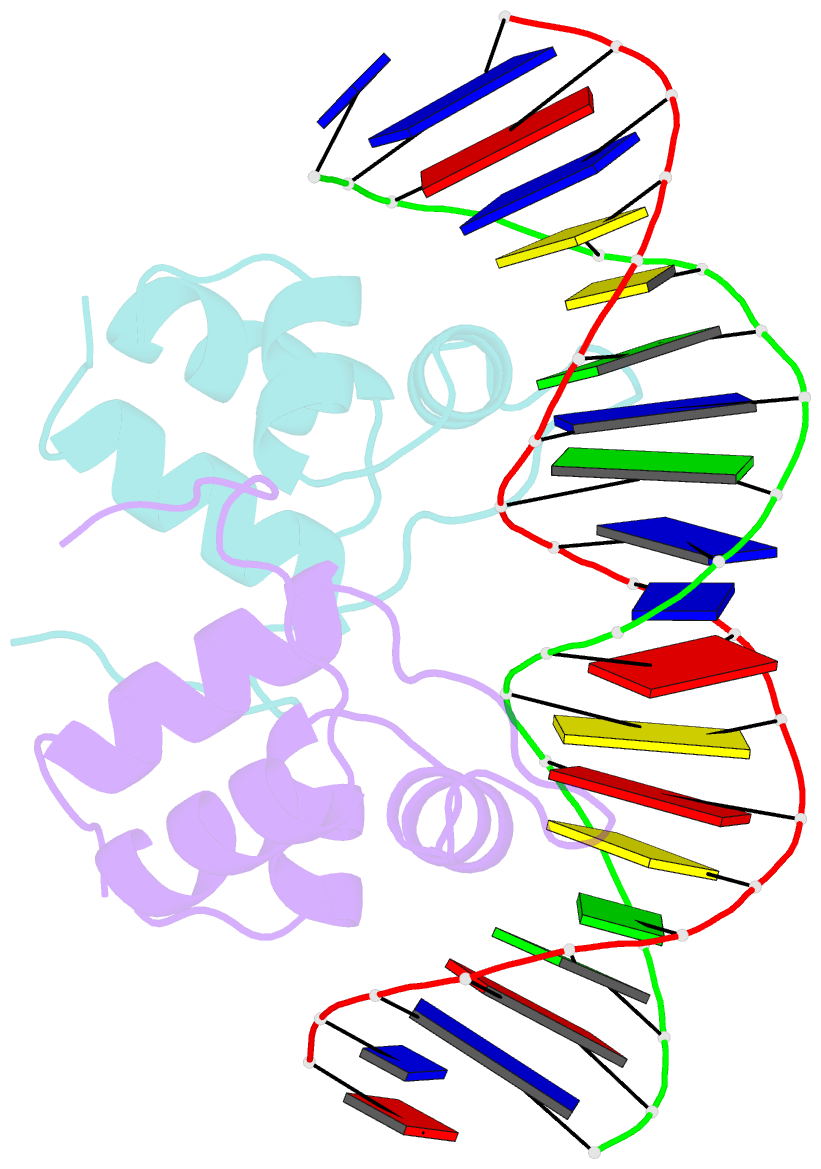
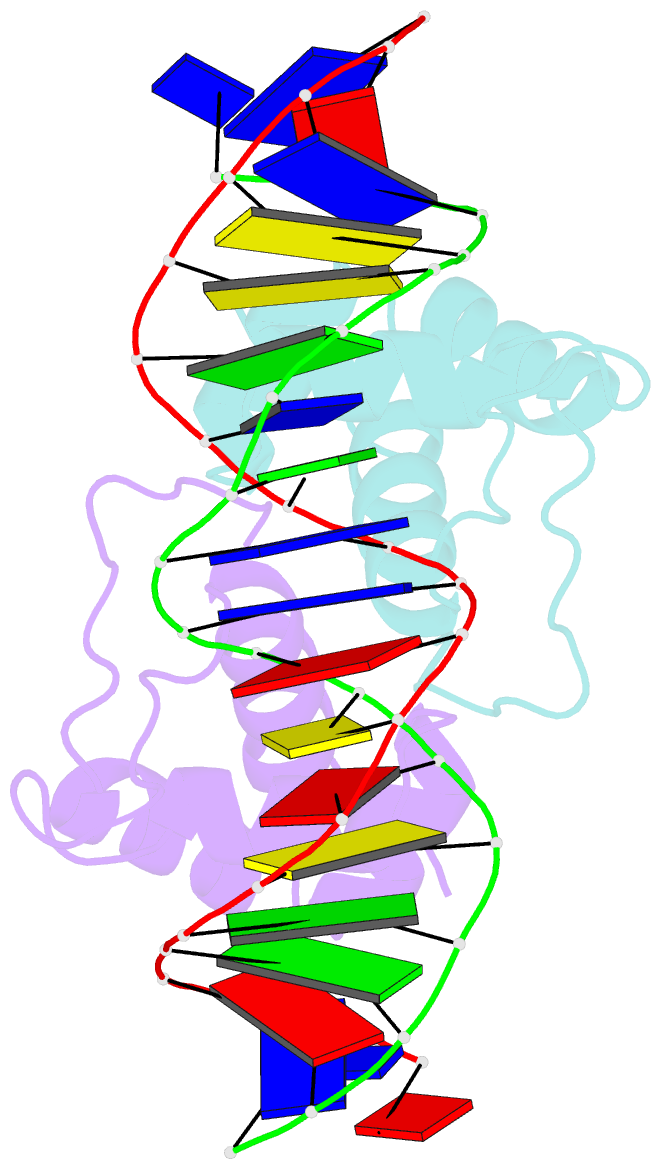
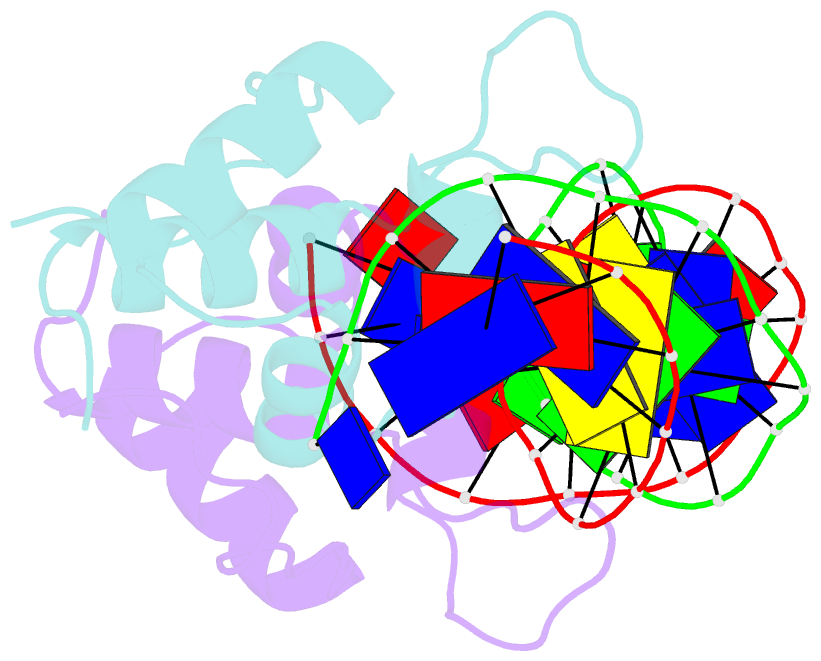
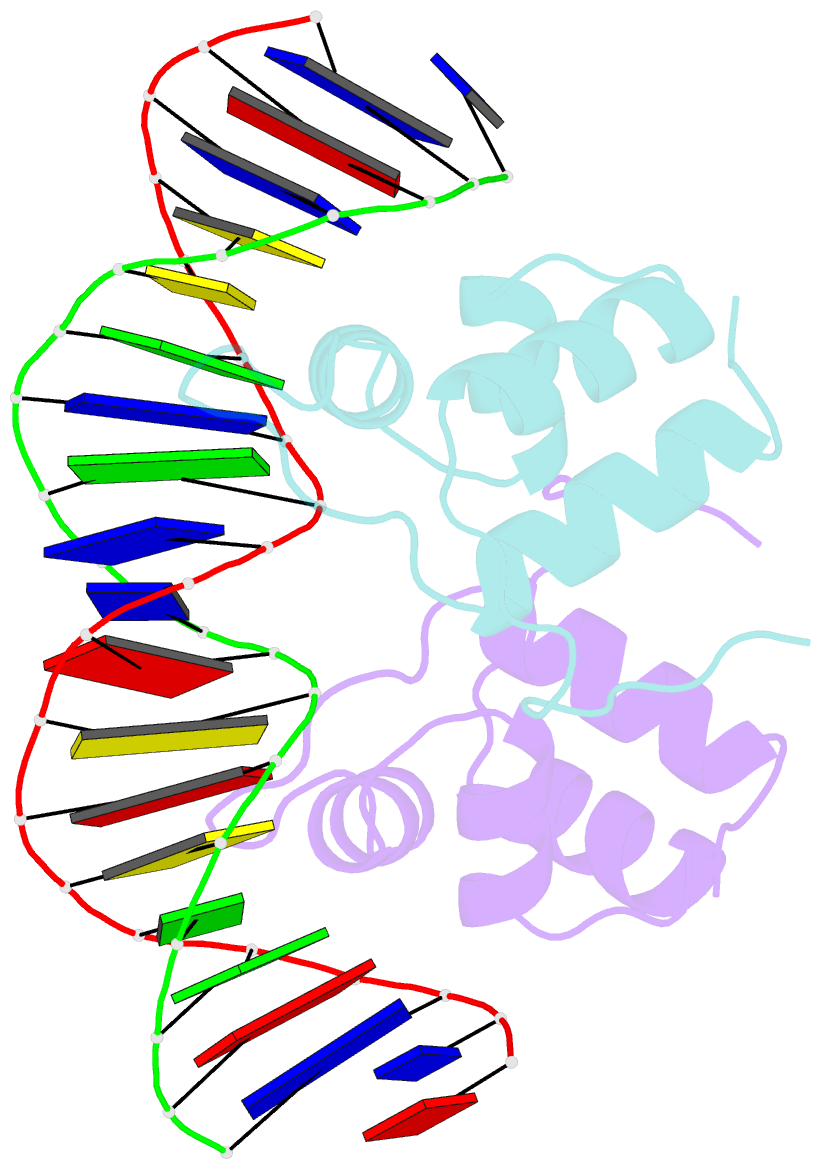
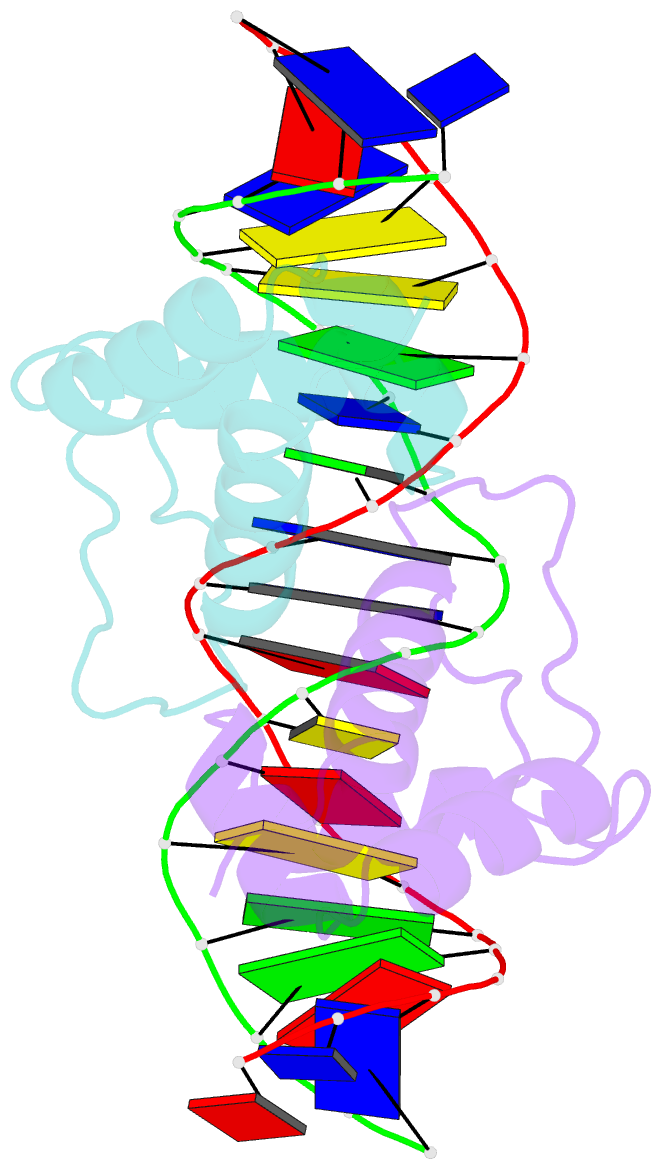
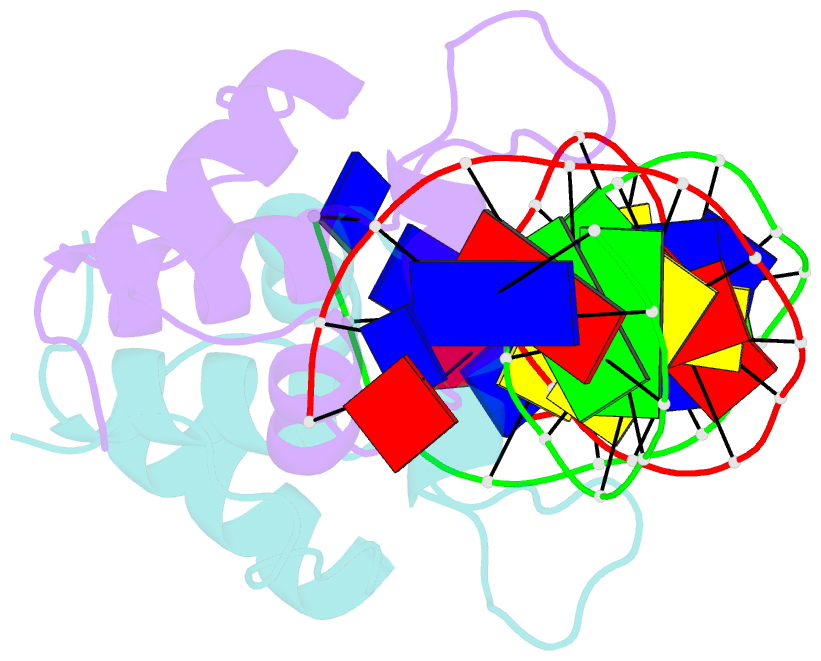
- Aca1 in complex with 19bp palindromic DNA substrate
- Reference
- Liu Y, Zhang L, Guo M, Chen L, Wu B, Huang H (2021): "Structural basis for anti-CRISPR repression mediated by bacterial operon proteins Aca1 and Aca2." J.Biol.Chem., 297, 101357. doi: 10.1016/j.jbc.2021.101357.
- Abstract
- It has been shown that phages have evolved anti-CRISPR (Acr) proteins to inhibit host CRISPR-Cas systems. Most acr genes are located upstream of anti-CRISPR-associated (aca) genes, which is instrumental for identifying these acr genes. Thus far, eight Aca families (Aca1-Aca8) have been identified, all proteins of which share low sequence homology and bind to different target DNA sequences. Recently, Aca1 and Aca2 proteins were discovered to function as repressors by binding to acr-aca promoters, thus implying a potential anti-anti-CRISPR mechanism. However, the structural basis for the repression roles of Aca proteins is still unknown. Here, we elucidated apo-structures of Aca1 and Aca2 proteins and their complex structures with their cognate operator DNA in two model systems, the Pseudomonas phage JBD30 and the Pectobacterium carotovorum template phage ZF40. In combination with biochemical and cellular assays, our study unveils dimerization and DNA-recognition mechanisms of Aca1 and Aca2 family proteins, thus revealing the molecular basis for Aca1-and Aca2-mediated anti-CRISPR repression. Our results also shed light on understanding the repression roles of other Aca family proteins and autoregulation roles of acr-aca operons.