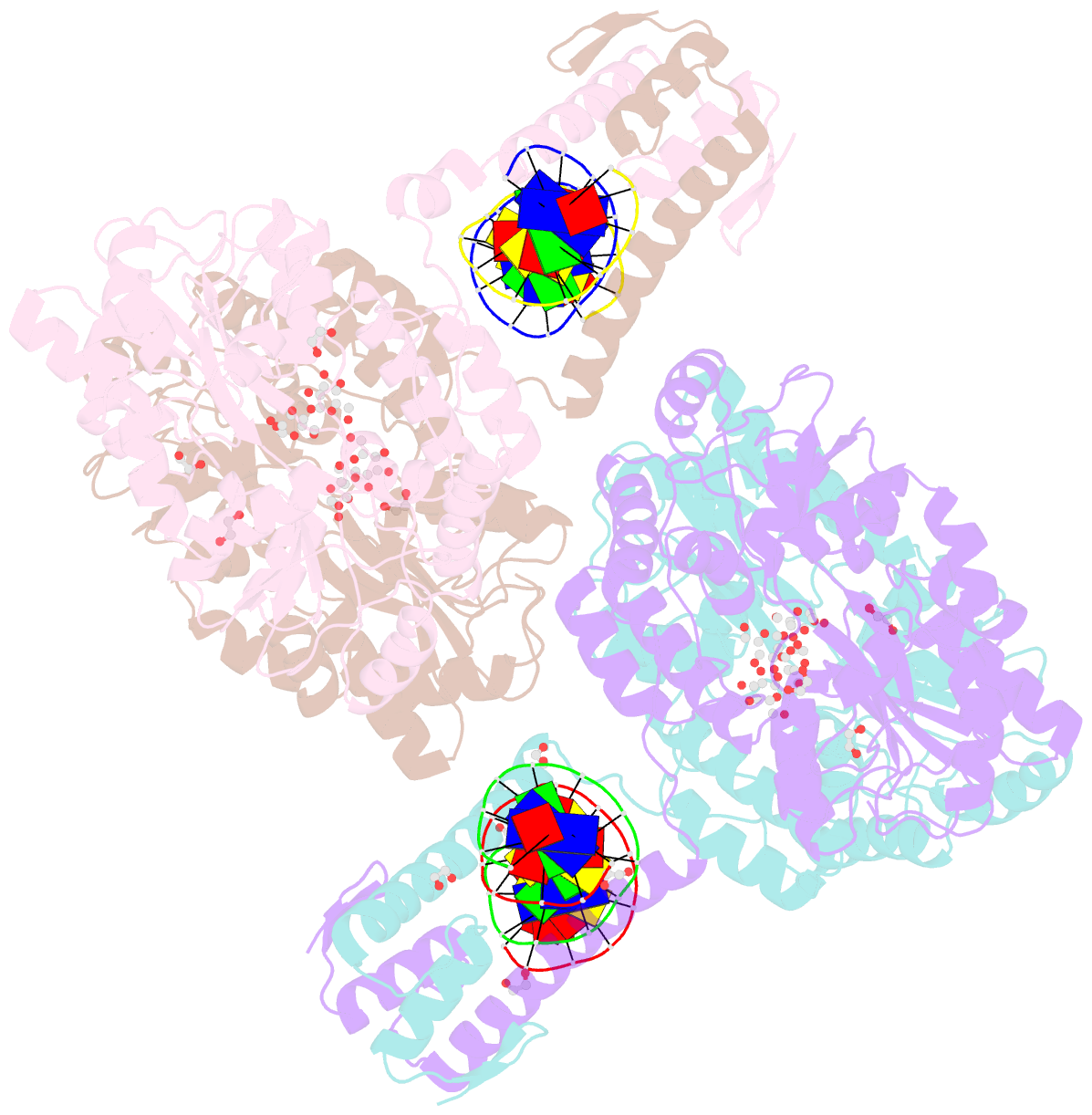
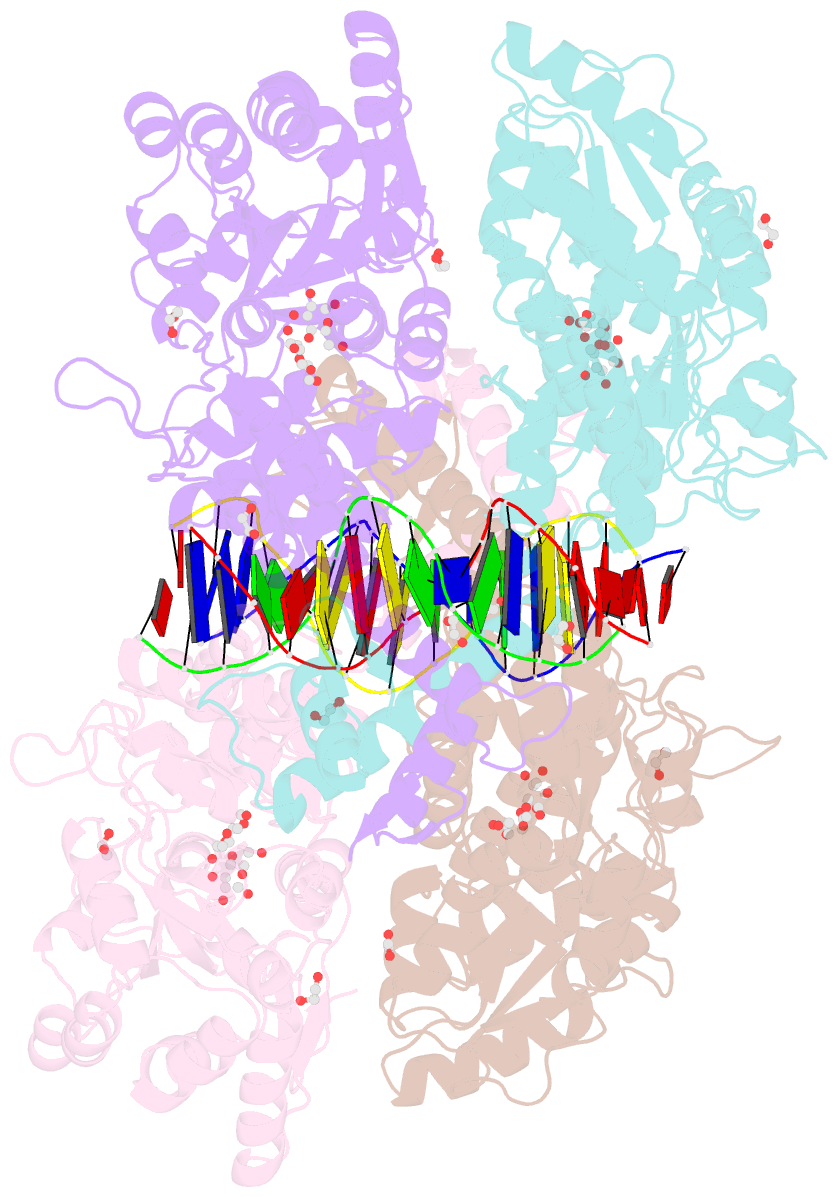
Summary information and primary citation
- PDB-id
- 7vn7; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription
- Method
- X-ray (2.11 Å)
- Summary
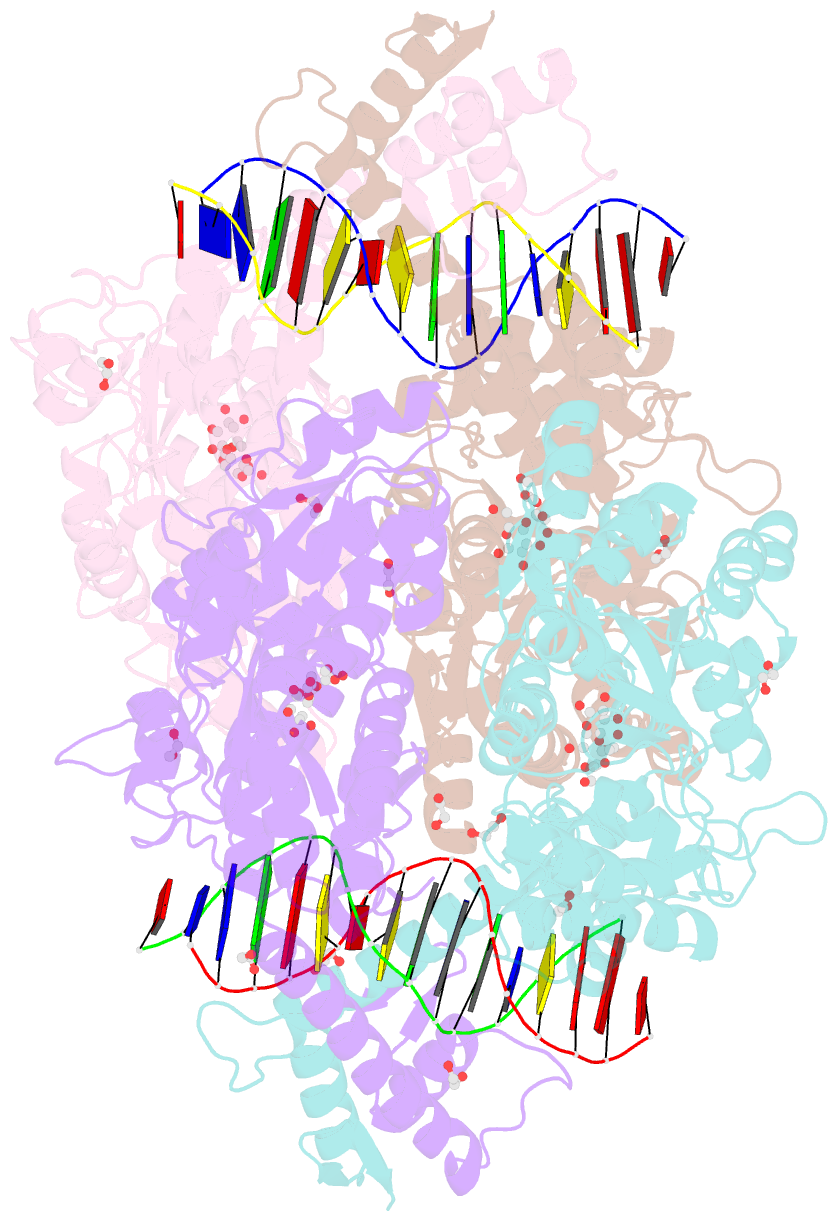
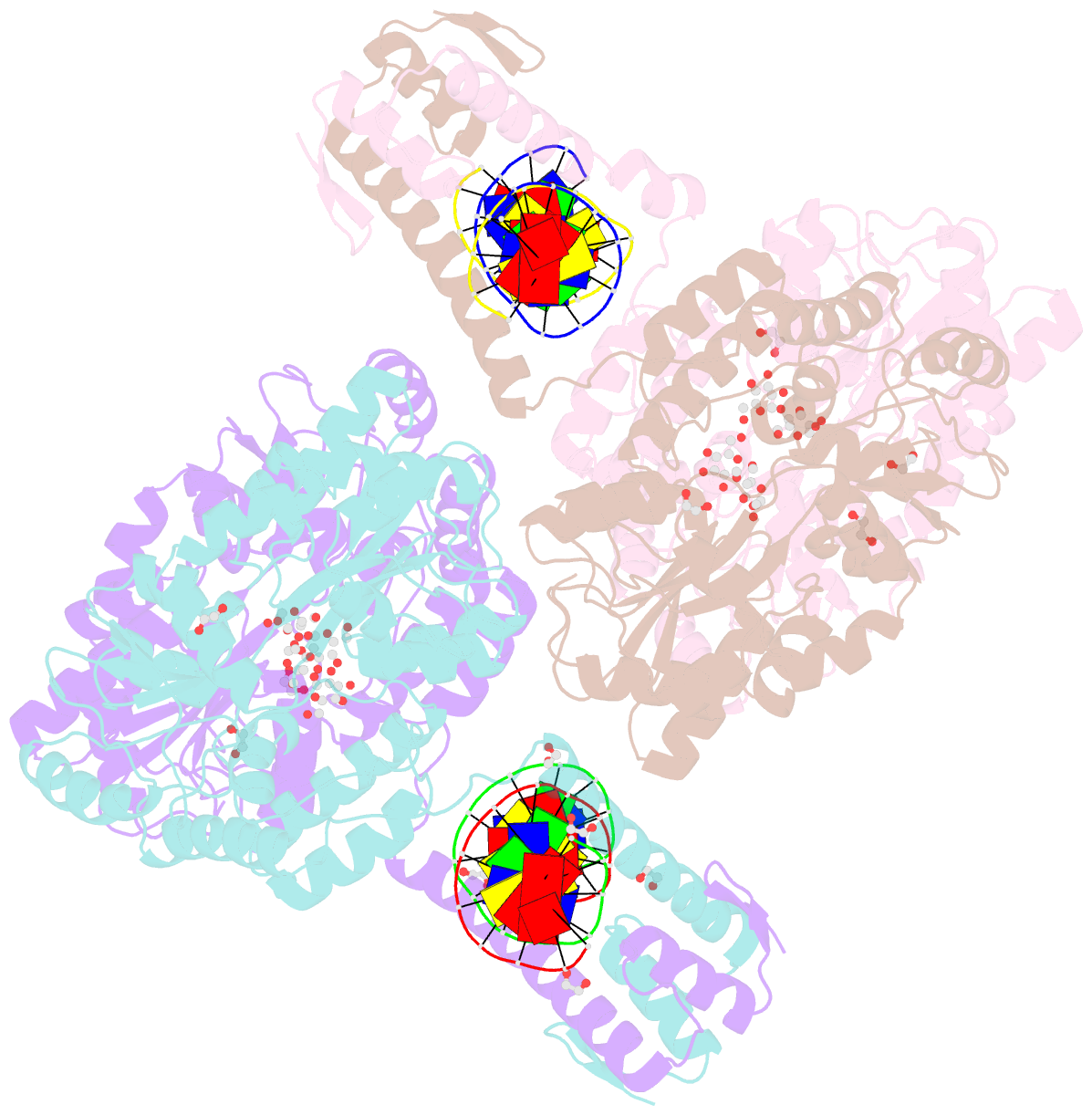
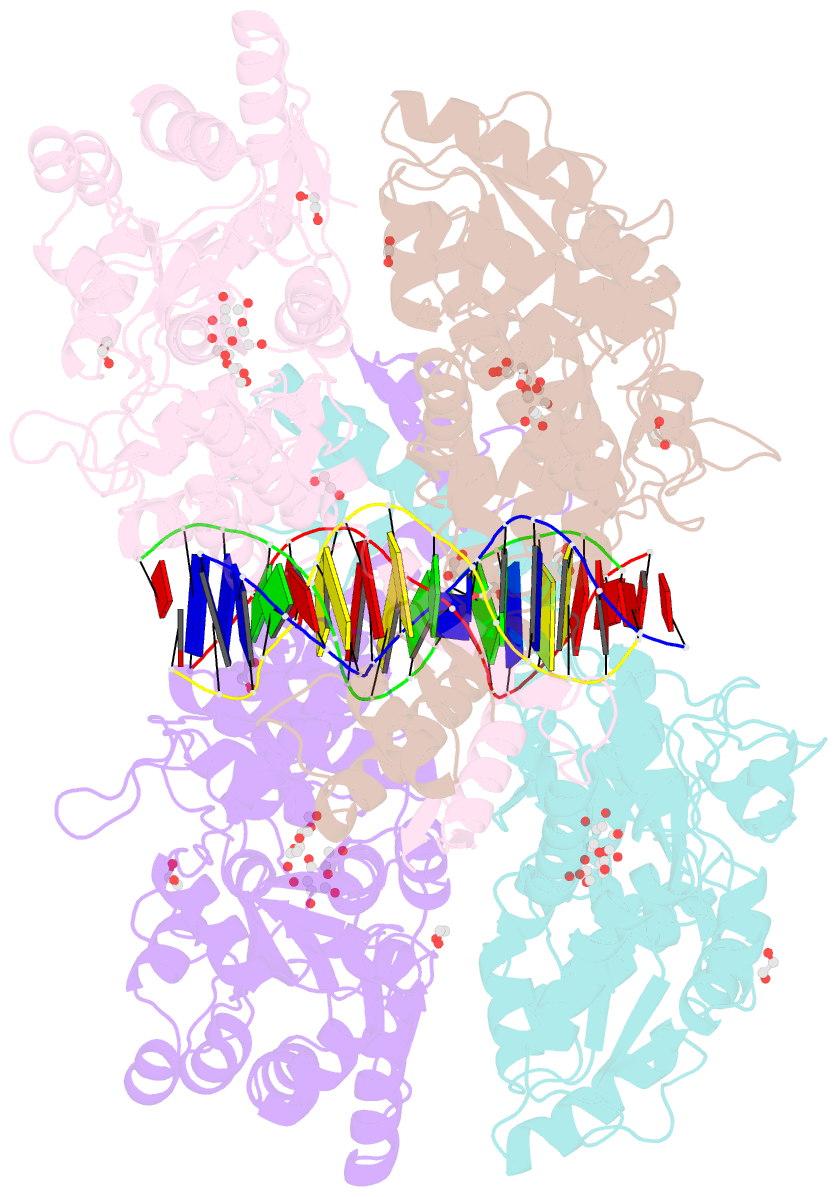
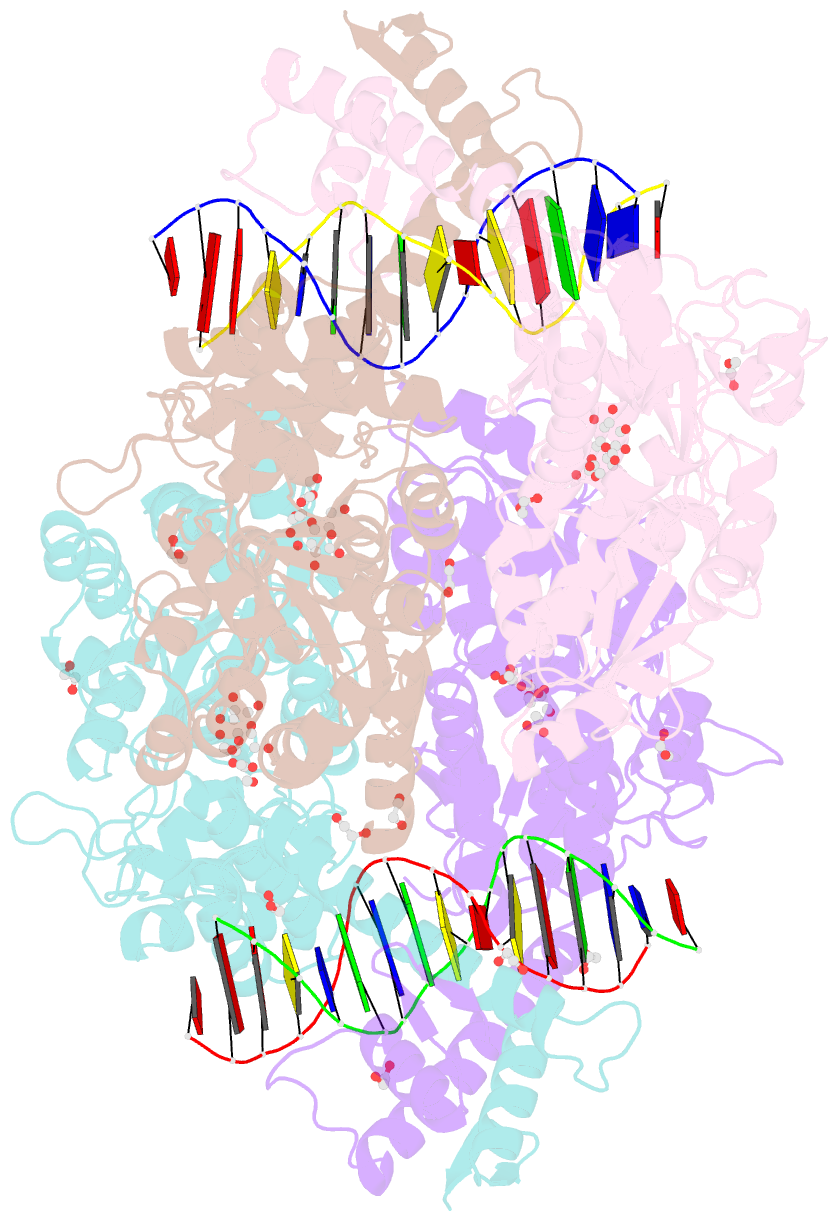
- Crystal structure of mbp-fused bil1-bzr1 (21-90) in complex with double-stranded DNA contaning gacacgtgtc
- Reference
- Nosaki S, Mitsuda N, Sakamoto S, Kusubayashi K, Yamagami A, Xu Y, Bui TBC, Terada T, Miura K, Nakano T, Tanokura M, Miyakawa T (2022): "Brassinosteroid-induced gene repression requires specific and tight promoter binding of BIL1/BZR1 via DNA shape readout." Nat.Plants, 8, 1440-1452. doi: 10.1038/s41477-022-01289-6.
- Abstract
- BRZ-INSENSITIVE-LONG 1 (BIL1)/BRASSINAZOLE-RESISTANT 1 (BZR1) and its homologues are plant-specific transcription factors that convert the signalling of the phytohormones brassinosteroids (BRs) to transcriptional responses, thus controlling various physiological processes in plants. Although BIL1/BZR1 upregulates some BR-responsive genes and downregulates others, the molecular mechanism underlying the dual roles of BIL1/BZR1 is still poorly understood. Here we show that BR-responsive transcriptional repression by BIL1/BZR1 requires the tight binding of BIL1/BZR1 alone to the 10 bp elements of DNA fragments containing the known 6 bp core-binding motifs at the centre. Furthermore, biochemical and structural evidence demonstrates that the selectivity for two nucleobases flanking the core motifs is realized by the DNA shape readout of BIL1/BZR1 without direct recognition of the nucleobases. These results elucidate the molecular and structural basis of transcriptional repression by BIL1/BZR1 and contribute to further understanding of the dual roles of BIL1/BZR1 in BR-responsive gene regulation.