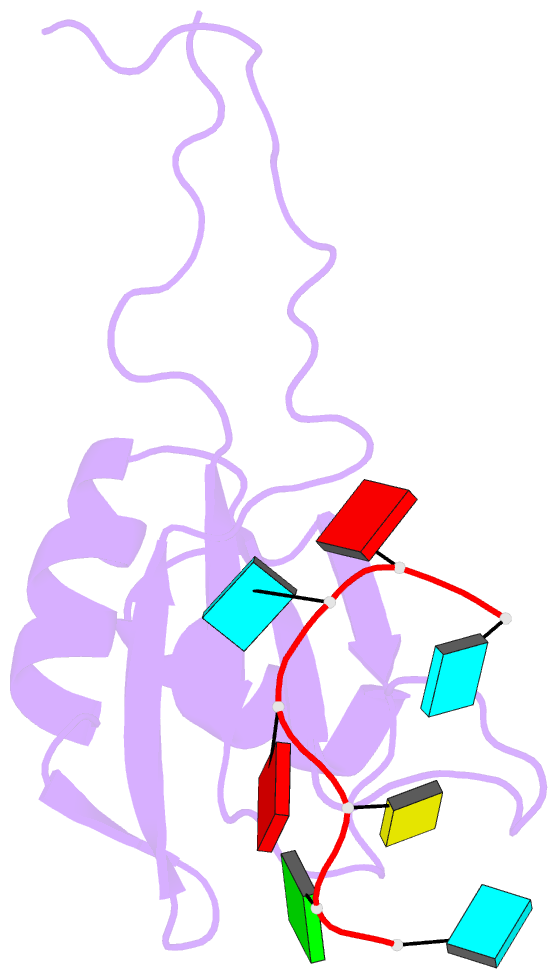
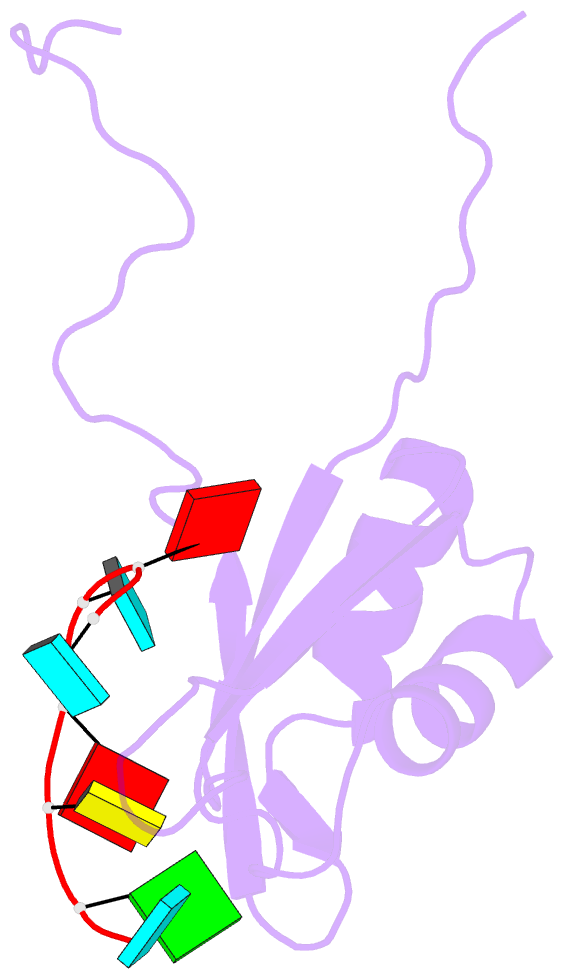
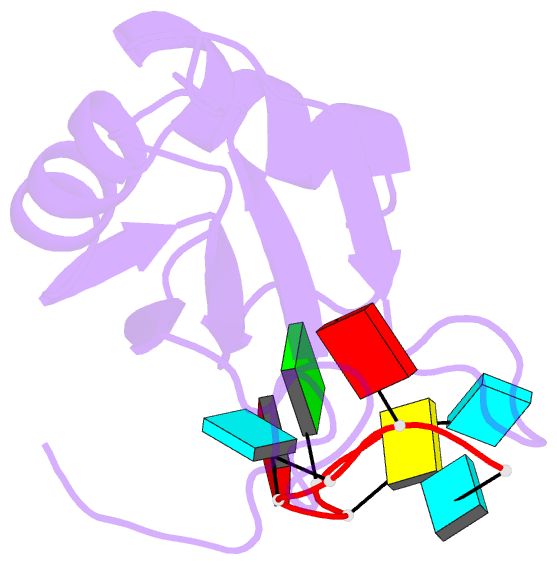
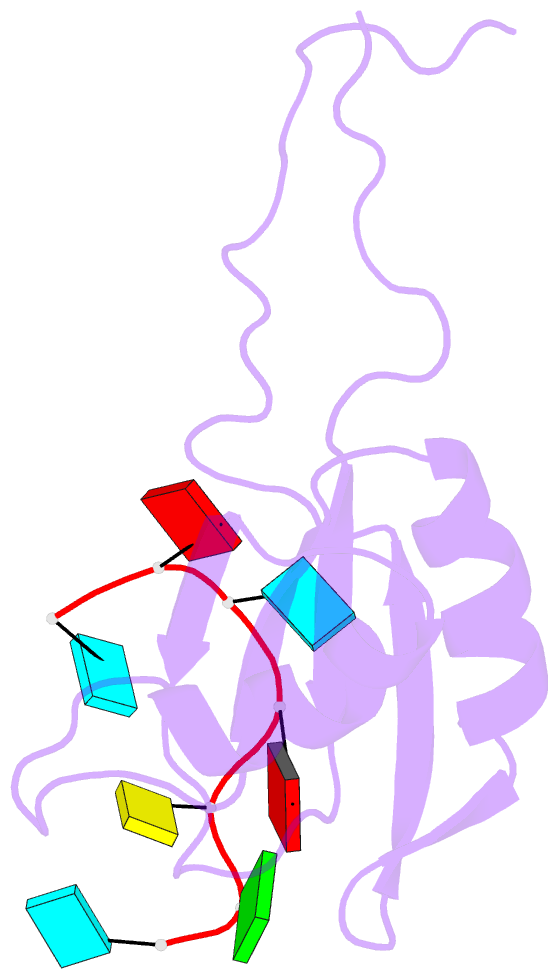
Summary information and primary citation
- PDB-id
- 7vrl; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- RNA binding protein-RNA
- Method
- NMR
- Summary
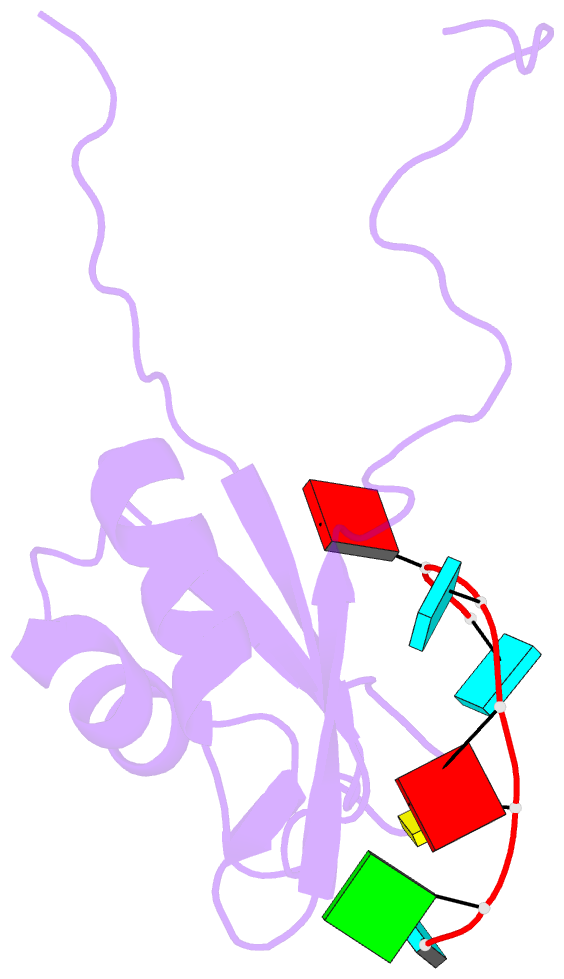
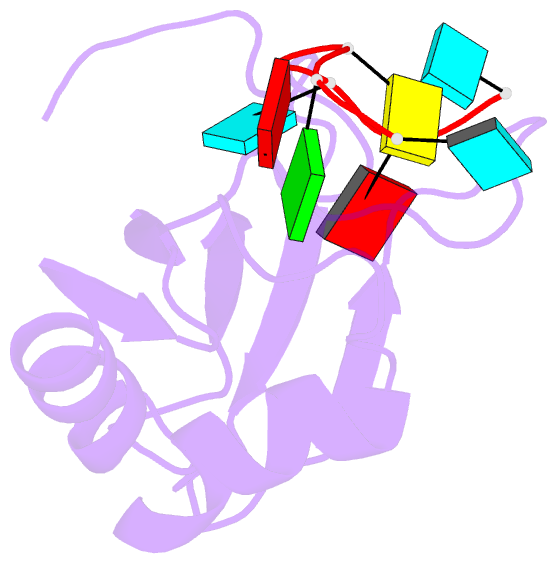
- Solution structure of rbfox rrm bound to a non-cognate RNA
- Reference
- Ye X, Yang W, Yi S, Zhao Y, Varani G, Jankowsky E, Yang F (2023): "Two distinct binding modes provide the RNA-binding protein RbFox with extraordinary sequence specificity." Nat Commun, 14, 701. doi: 10.1038/s41467-023-36394-3.
- Abstract
- Specificity of RNA-binding proteins for target sequences varies considerably. Yet, it is not understood how certain few proteins achieve markedly higher sequence specificity than most others. Here we show that the RNA Recognition Motif of RbFox accomplishes extraordinary sequence specificity by employing functionally and structurally distinct binding modes. Affinity measurements of RbFox for all binding site variants reveal the existence of two distinct binding modes. The first exclusively accommodates cognate and closely related RNAs with high affinity. The second mode accommodates all other RNAs with reduced affinity by imposing large thermodynamic penalties on non-cognate sequences. NMR studies indicate marked structural differences between the two binding modes, including large conformational rearrangements distant from the RNA-binding site. Distinct binding modes by a single RNA-binding module explain extraordinary sequence selectivity and reveal an unknown layer of functional diversity, cross talk and regulation in RNA-protein interactions.