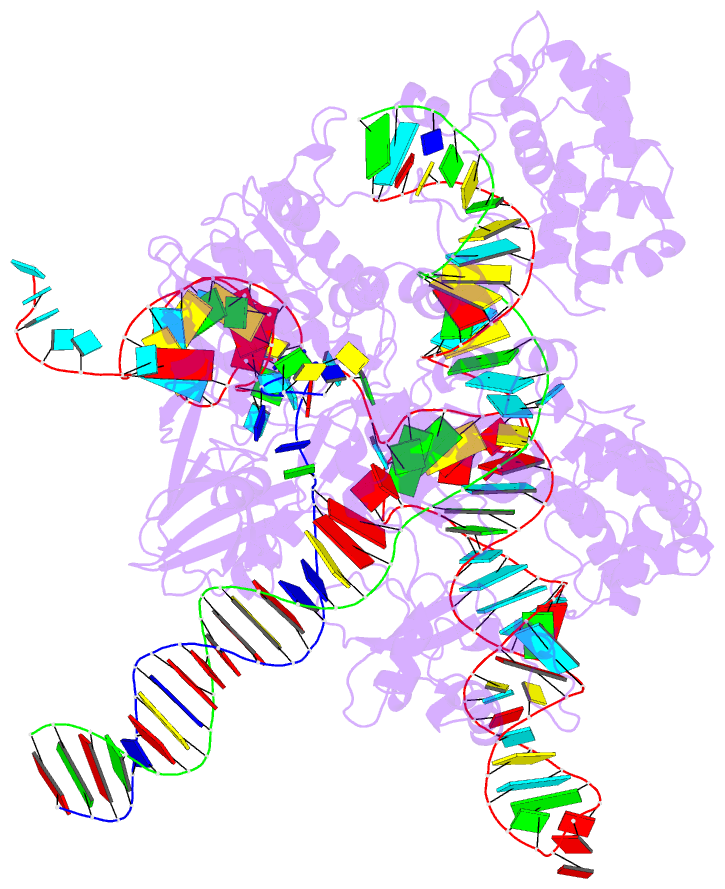
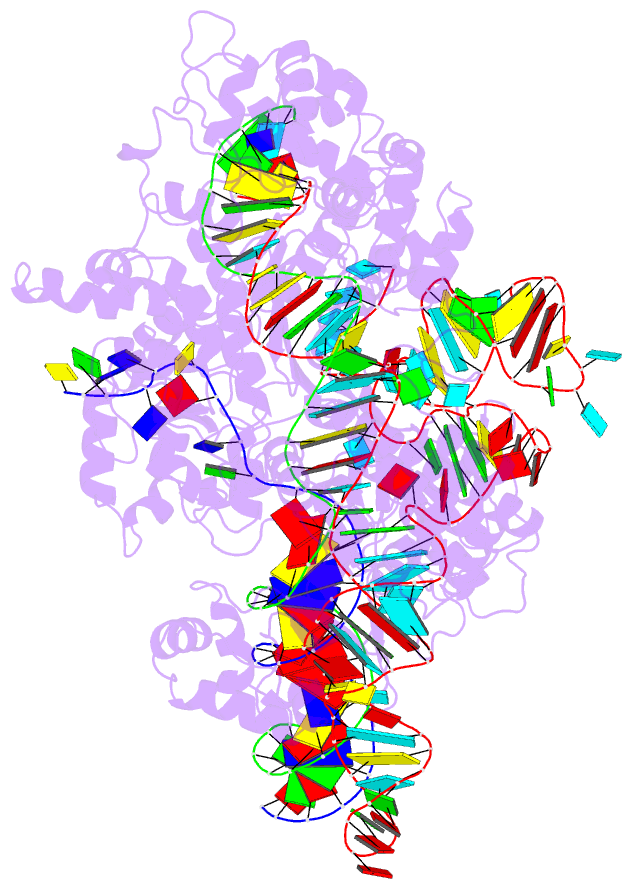
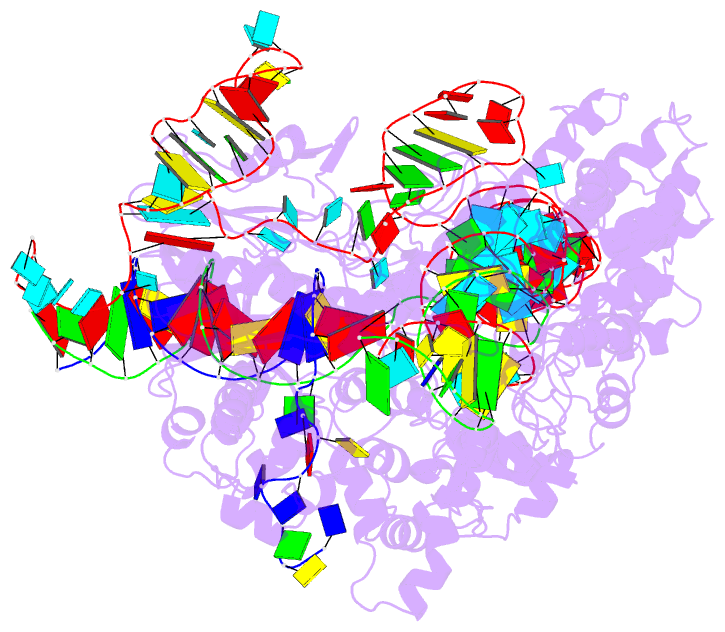
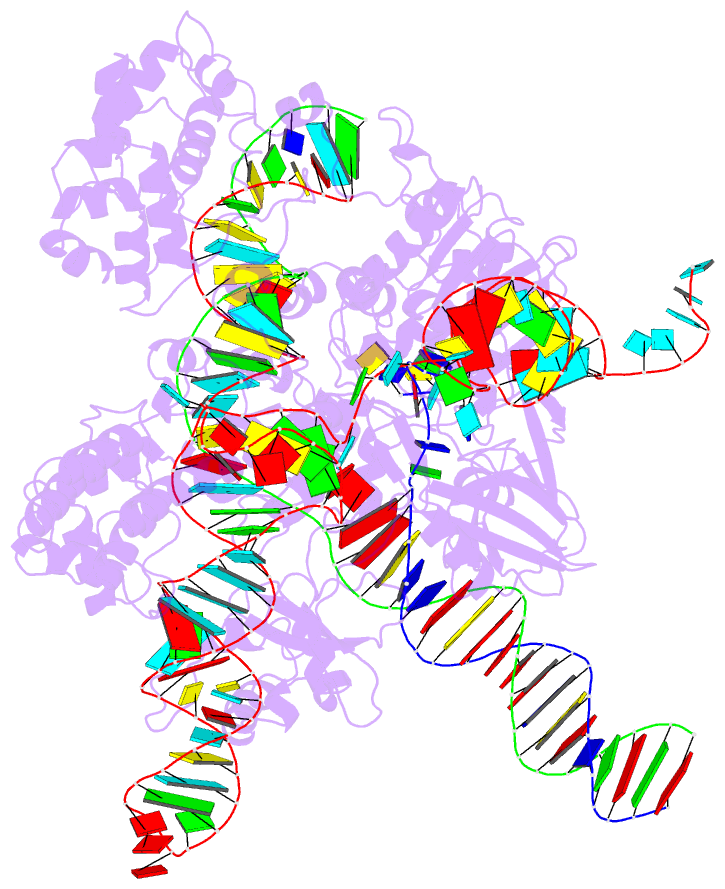
Summary information and primary citation
- PDB-id
- 7vw3; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- DNA binding protein-RNA-DNA
- Method
- cryo-EM (3.8 Å)
- Summary
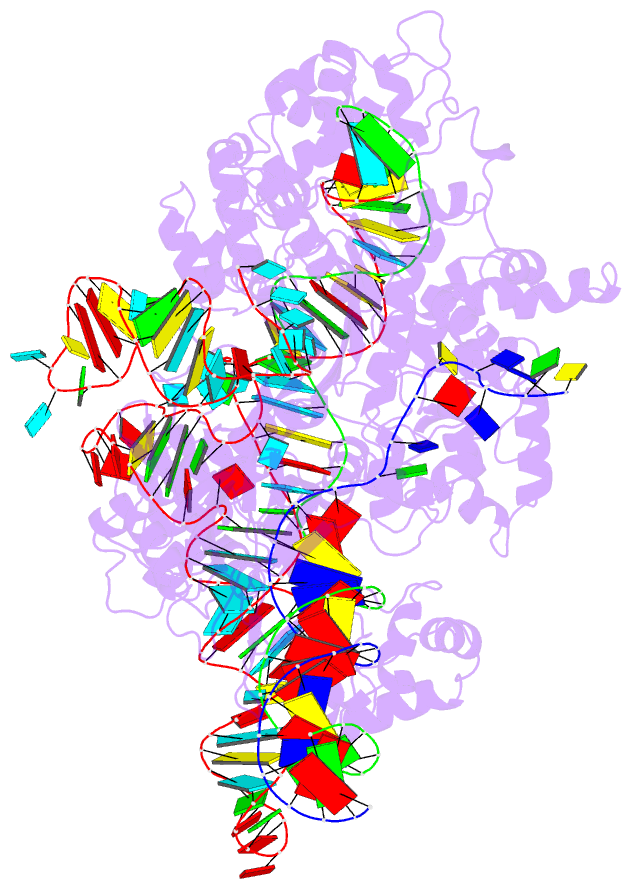
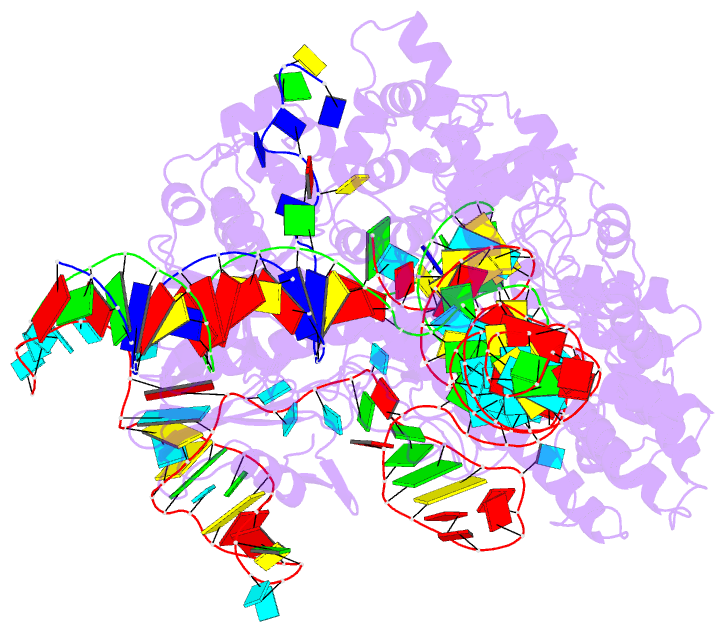
- cryo-EM structure of sacas9-sgrna-DNA ternary complex
- Reference
- Du W, Zhu H, Qian J, Xue D, Zheng S, Huang Q (2023): "Full-Length Model of SaCas9-sgRNA-DNA Complex in Cleavage State." Int J Mol Sci, 24. doi: 10.3390/ijms24021204.
- Abstract
- Staphylococcus aureus Cas9 (SaCas9) is a widely used genome editing tool. Understanding its molecular mechanisms of DNA cleavage could effectively guide the engineering optimization of this system. Here, we determined the first cryo-electron microscopy structure of the SaCas9-sgRNA-DNA ternary complex. This structure reveals that the HNH nuclease domain is tightly bound to the cleavage site of the target DNA strand, and is in close contact with the WED and REC domains. Moreover, it captures the complete structure of the sgRNA, including the previously unresolved stem-loop 2. Based on this structure, we build a full-length model for the ternary complex in cleavage state. This model enables identification of the residues for the interactions between the HNH domain and the WED and REC domains. Moreover, we found that the stem-loop 2 of the sgRNA tightly binds to the PI and RuvC domains and may also regulate the position shift of the RuvC domain. Further mutagenesis and molecular dynamics simulations supported the idea that the interactions of the HNH domain with the WED and REC domains play an important role in the DNA cleavage. Thus, this study provides new mechanistic insights into the DNA cleavage of SaCas9 and is also useful for guiding the future engineering of SaCas9-mediated gene editing systems.