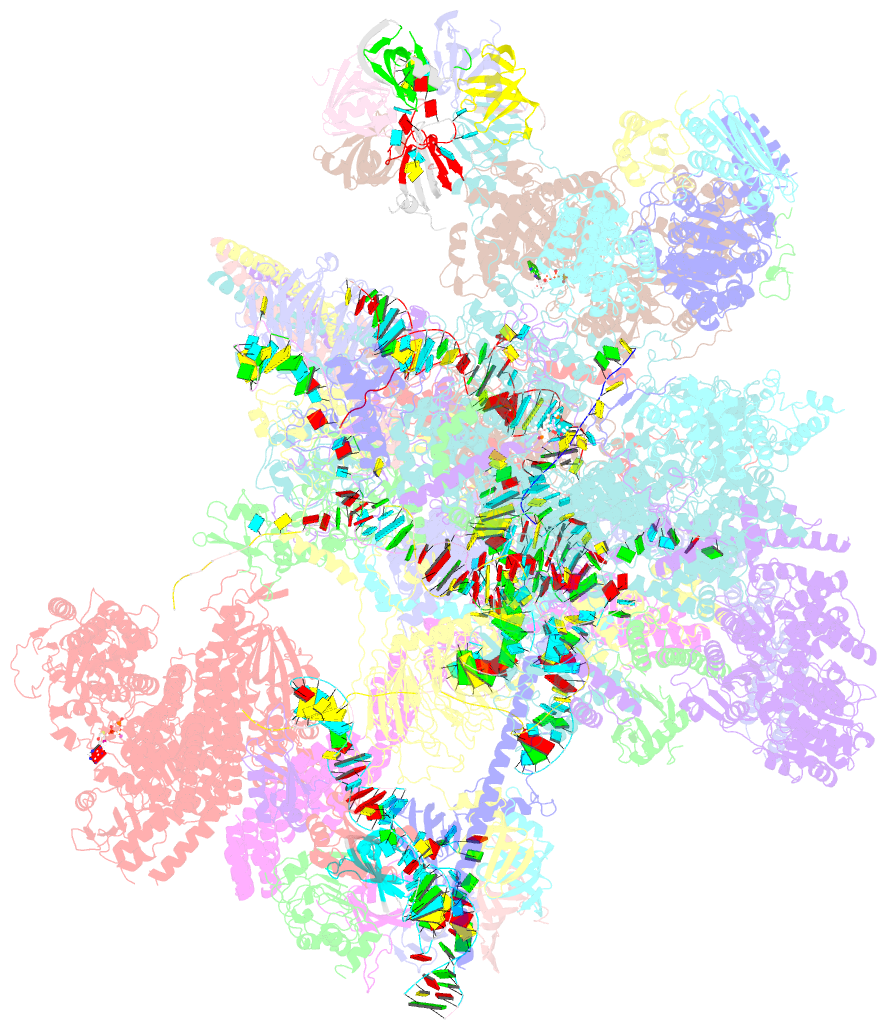
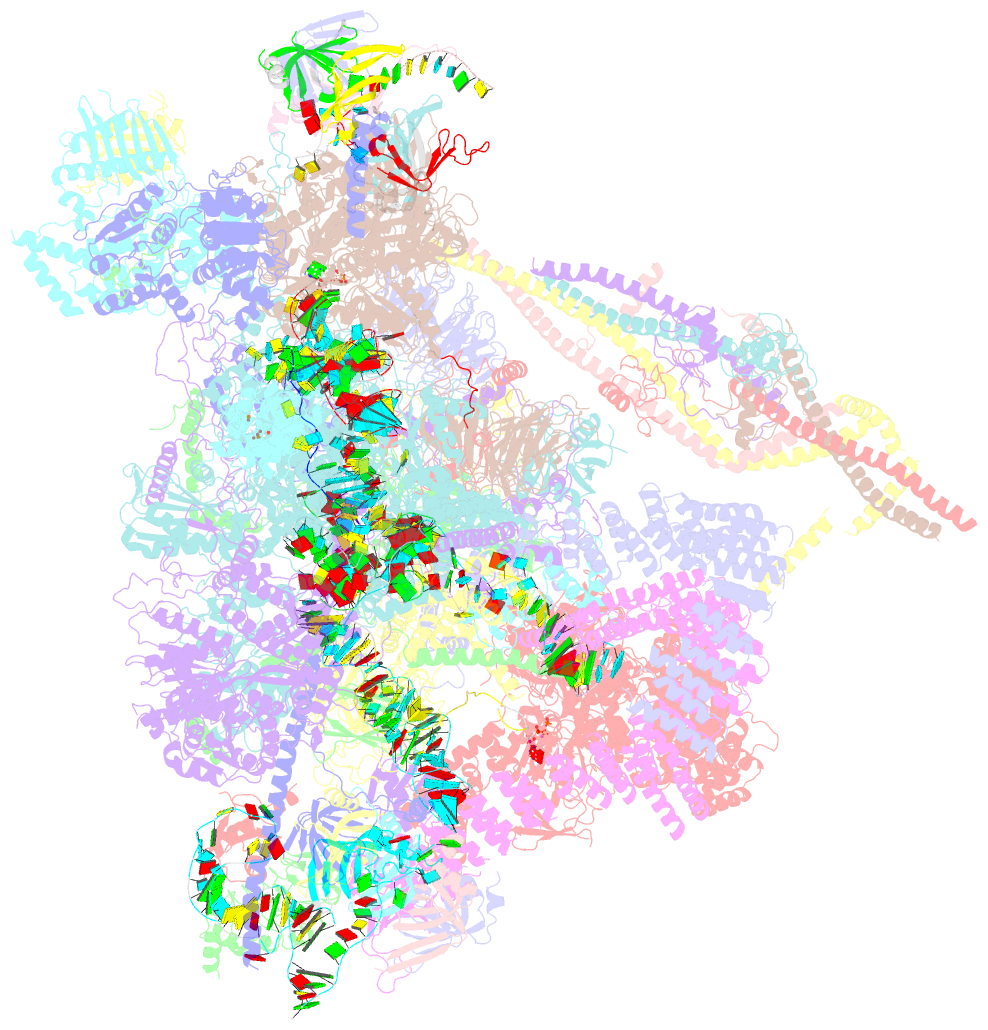
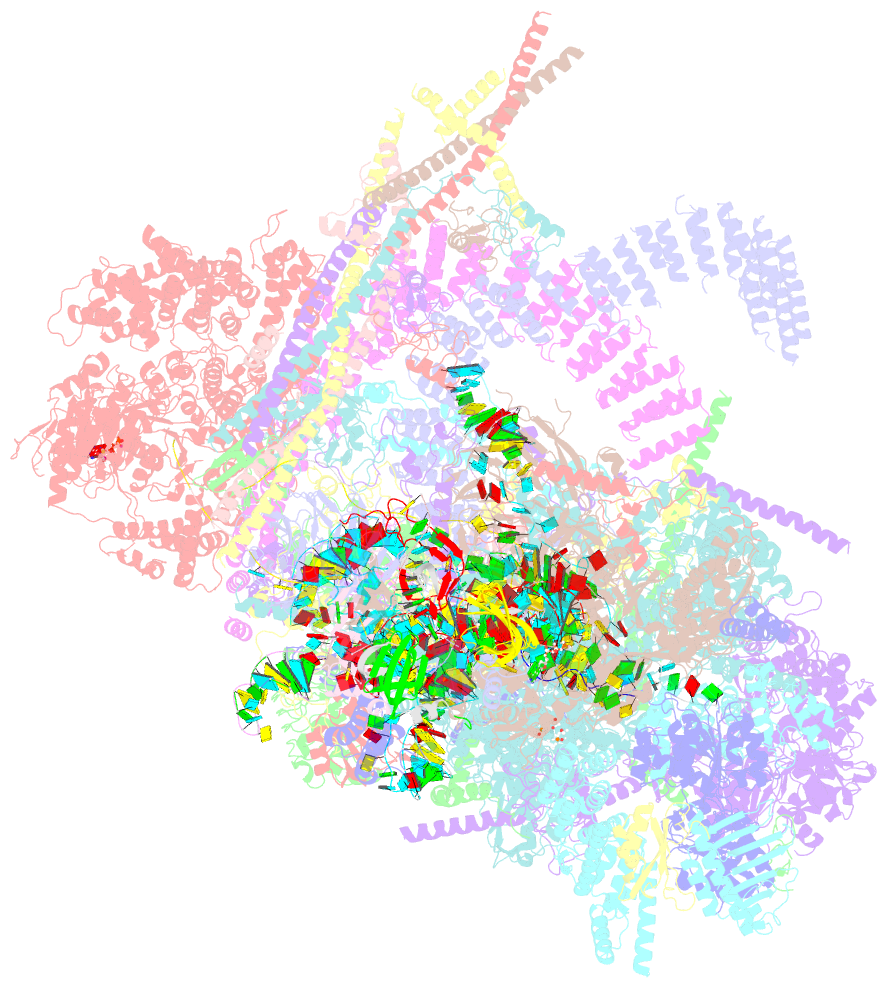
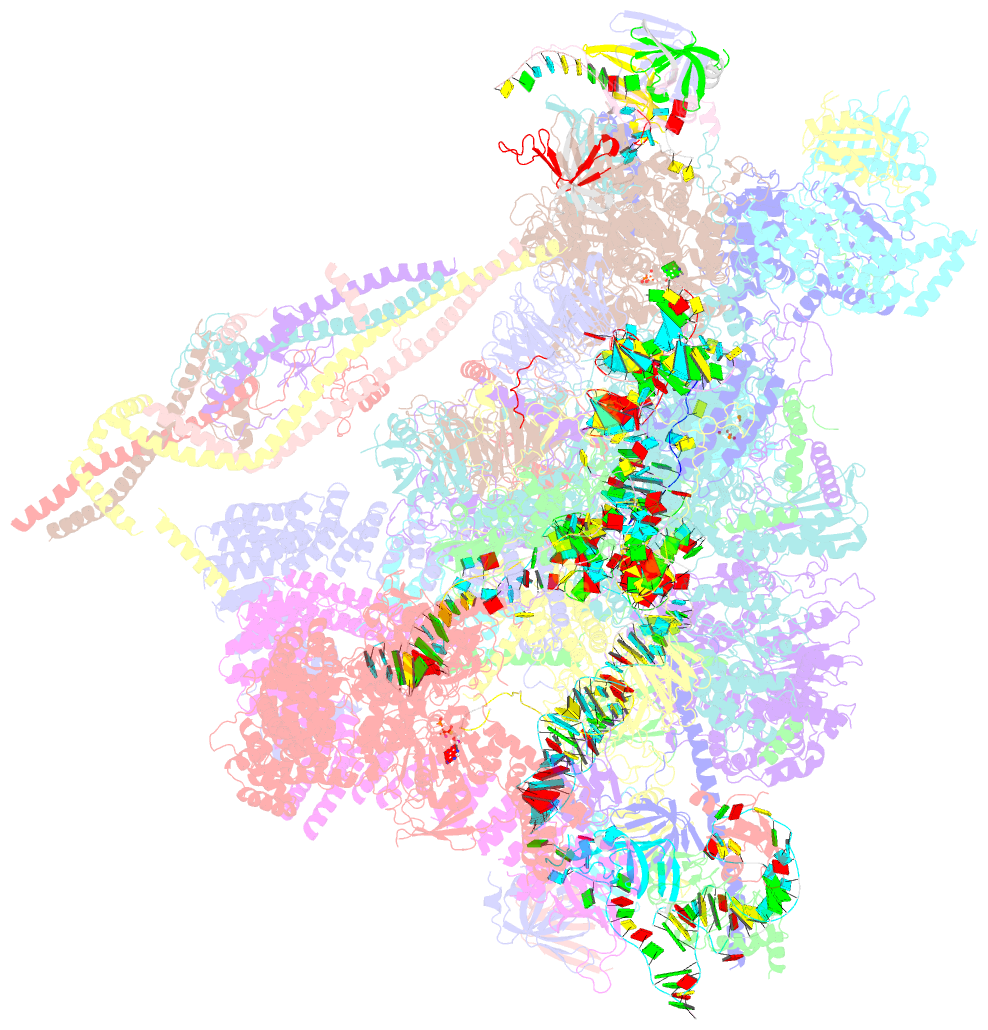
Summary information and primary citation
- PDB-id
- 7w5a; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- splicing
- Method
- cryo-EM (3.6 Å)
- Summary
- The cryo-EM structure of human pre-c*-ii complex
- Reference
- Zhan X, Lu Y, Zhang X, Yan C, Shi Y (2022): "Mechanism of exon ligation by human spliceosome." Mol.Cell, 82, 2769-2778.e4. doi: 10.1016/j.molcel.2022.05.021.
- Abstract
- Pre-mRNA splicing involves two sequential reactions: branching and exon ligation. The C complex after branching undergoes remodeling to become the C∗ complex, which executes exon ligation. Here, we report cryo-EM structures of two intermediate human spliceosomal complexes, pre-C∗-I and pre-C∗-II, both at 3.6 Å. In both structures, the 3' splice site is already docked into the active site, the ensuing 3' exon sequences are anchored on PRP8, and the step II factor FAM192A contacts the duplex between U2 snRNA and the branch site. In the transition of pre-C∗-I to pre-C∗-II, the step II factors Cactin, FAM32A, PRKRIP1, and SLU7 are recruited. Notably, the RNA helicase PRP22 is positioned quite differently in the pre-C∗-I, pre-C∗-II, and C∗ complexes, suggesting a role in 3' exon binding and proofreading. Together with information on human C and C∗ complexes, our studies recapitulate a molecular choreography of the C-to-C∗ transition, revealing mechanistic insights into exon ligation.