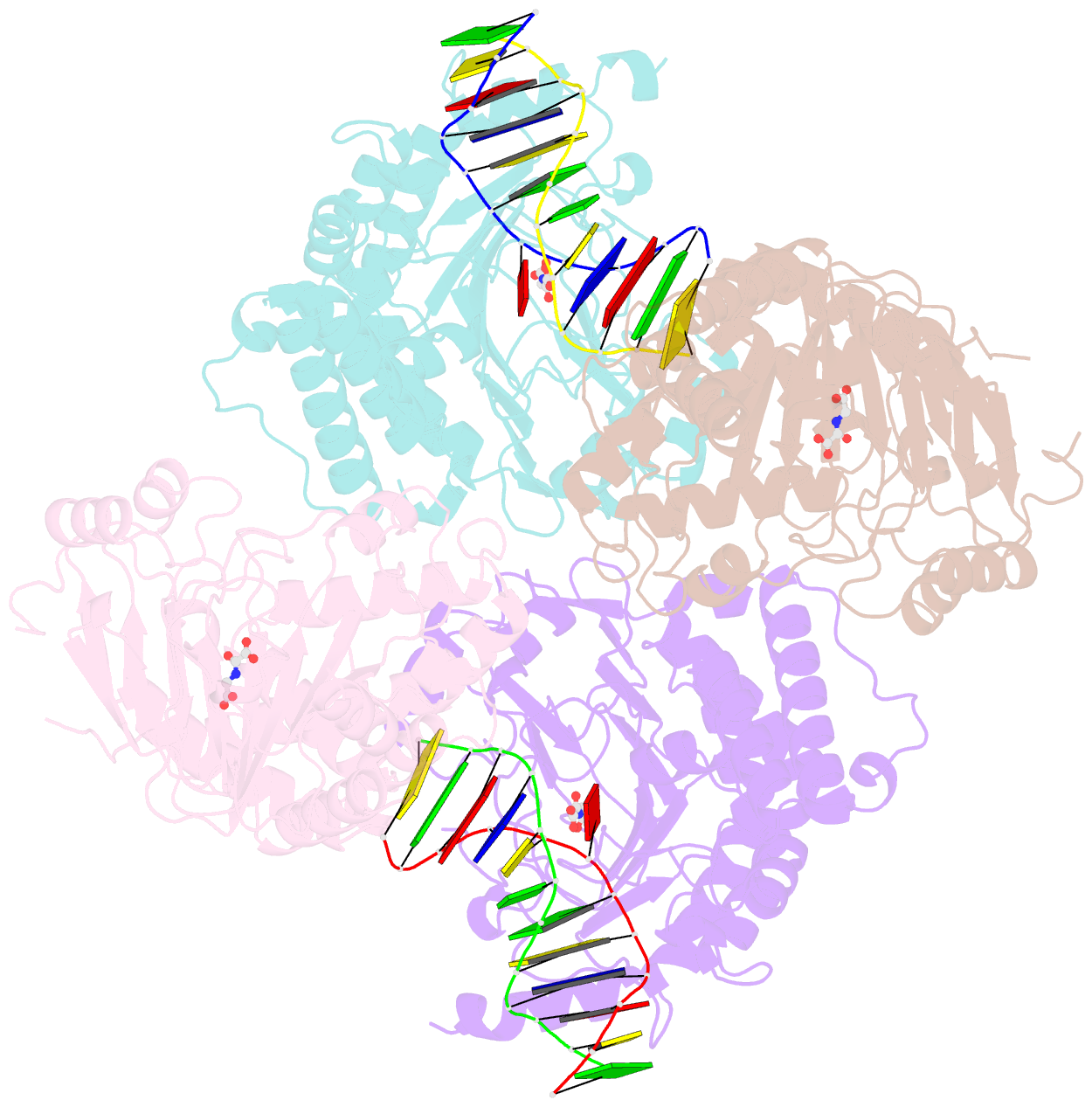
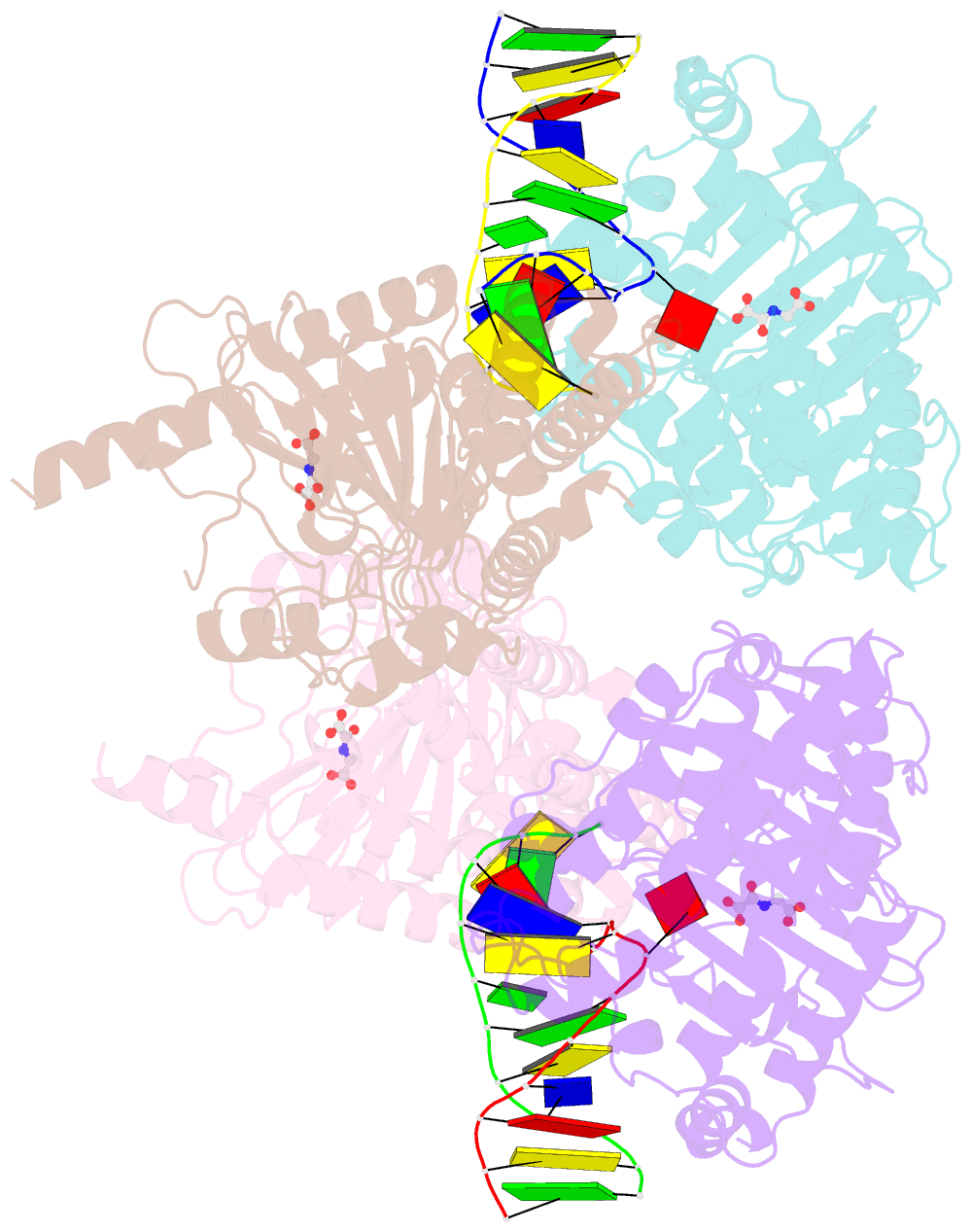
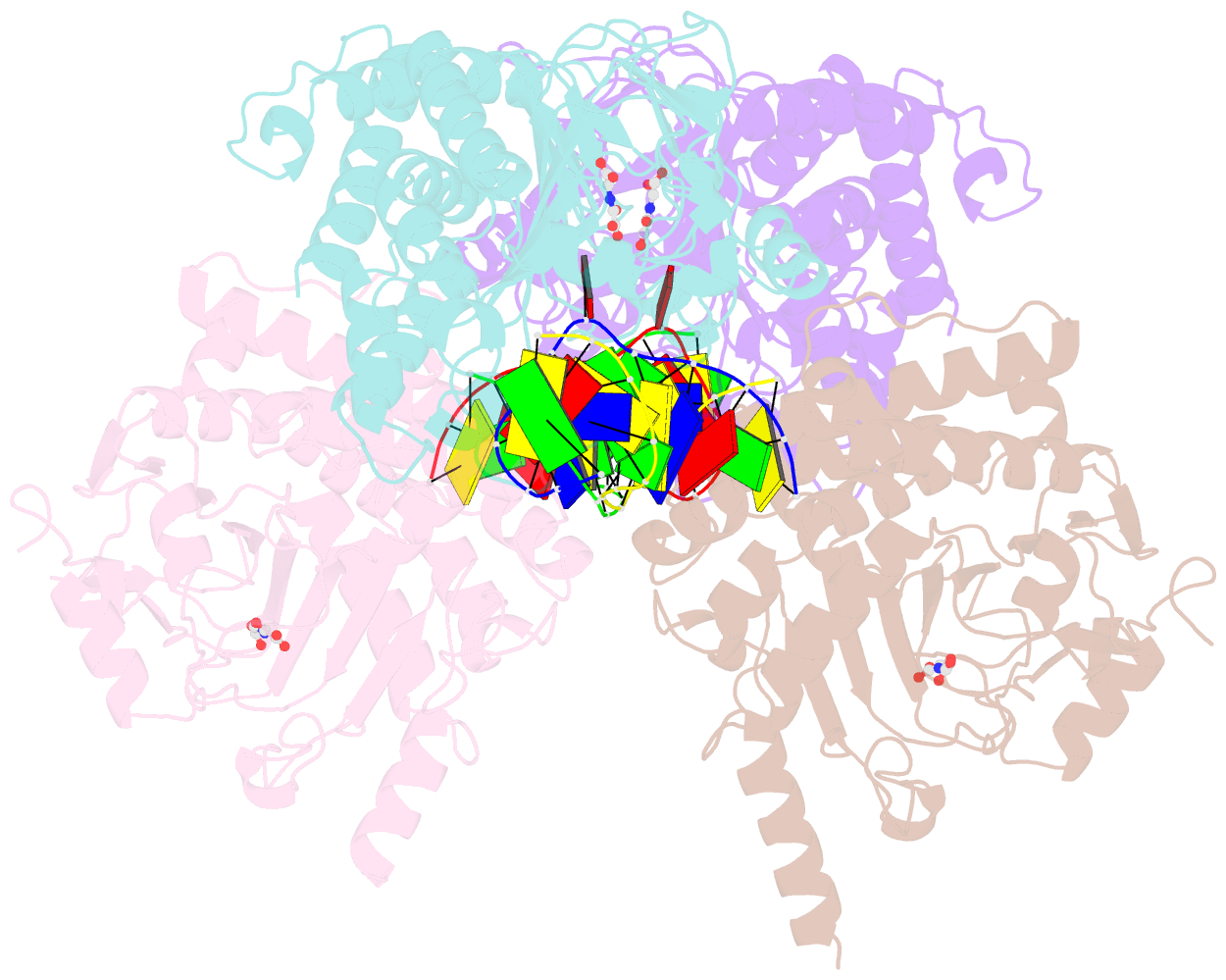
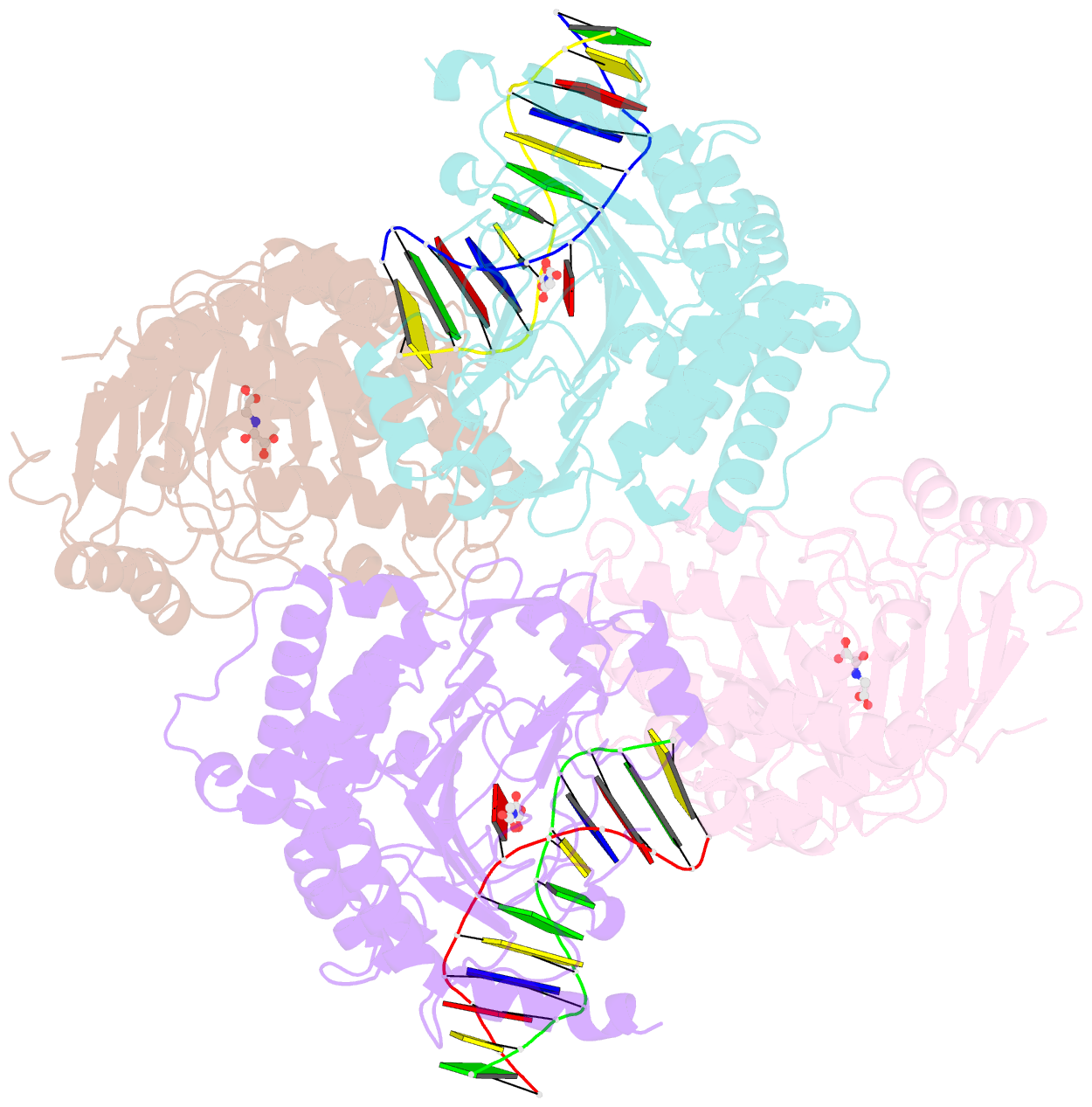
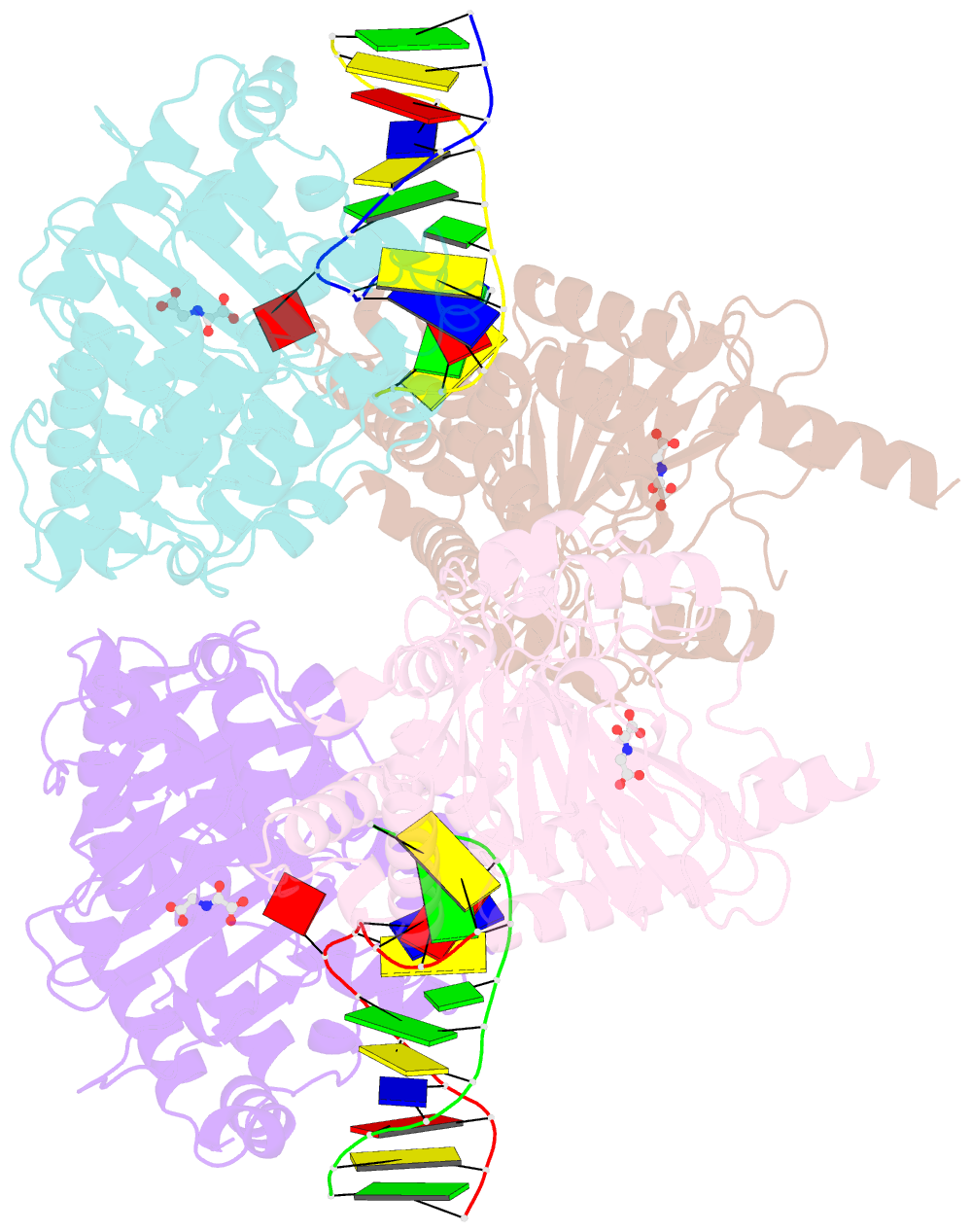
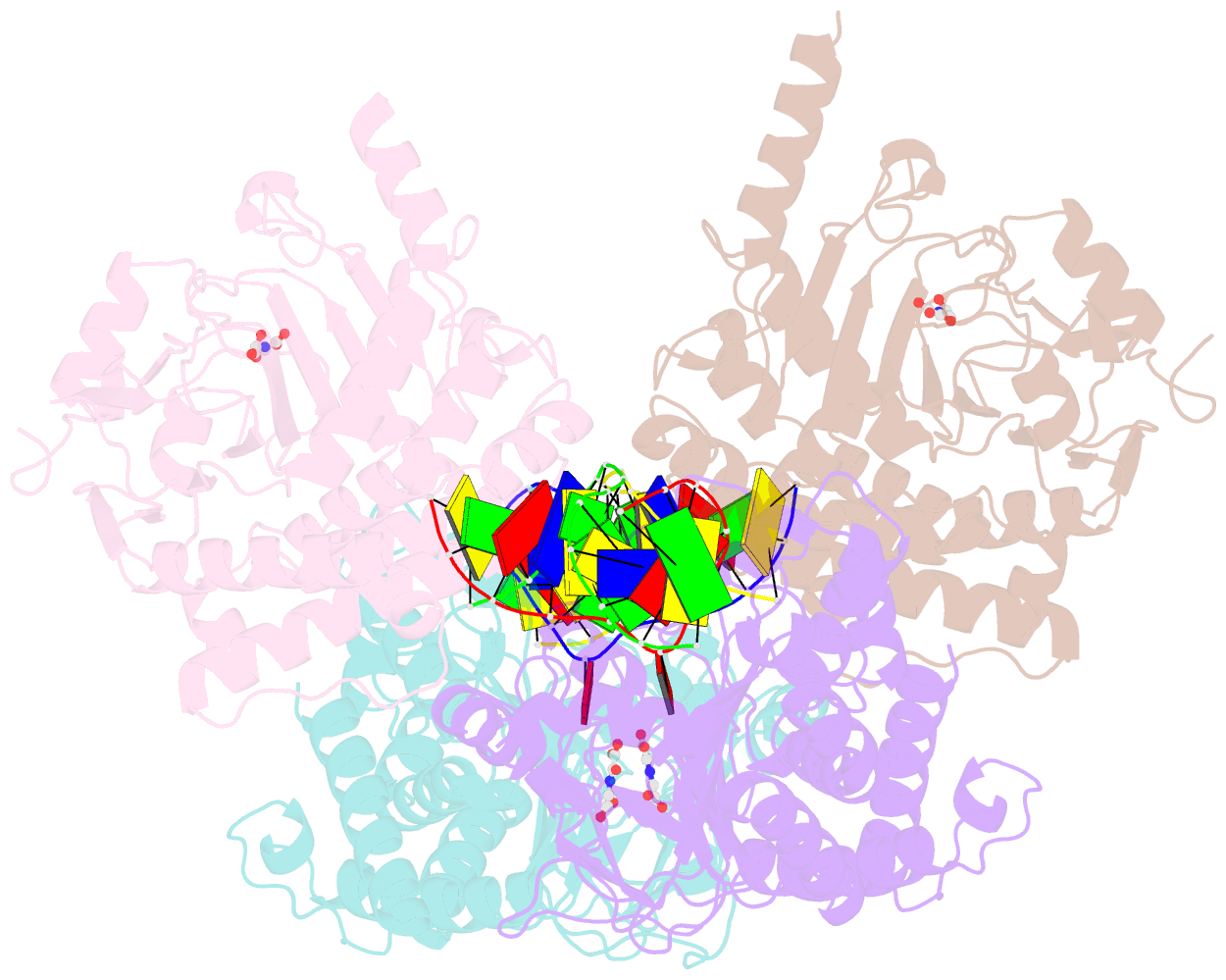
Summary information and primary citation
- PDB-id
- 7w5p; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- DNA binding protein-DNA
- Method
- X-ray (2.3 Å)
- Summary
- Crystal structure of the dioxygenase cctet from coprinopsis cinereain bound to 12bp n6-methyldeoxyadenine (6ma) containing duplex DNA
- Reference
- Mu Y, Zhang L, Hu J, Zhou J, Lin HW, He C, Chen HZ, Zhang L (2022): "A fungal dioxygenase CcTet serves as a eukaryotic 6mA demethylase on duplex DNA." Nat.Chem.Biol., 18, 733-741. doi: 10.1038/s41589-022-01041-3.
- Abstract
- N6-methyladenosine (6mA) is a DNA modification that has recently been found to play regulatory roles during mammalian early embryo development and mitochondrial transcription. We found that a dioxygenase CcTet from the fungus Coprinopsis cinerea is also a dsDNA 6mA demethylase. It oxidizes 6mA to the intermediate N6-hydroxymethyladenosine (6hmA) with robust activity of 6mA-containing duplex DNA (dsDNA) as well as isolated genomics DNA. Structural characterization revealed that CcTet utilizes three flexible loop regions and two key residues-D337 and G331-in the active pocket to preferentially recognize substrates on dsDNA. A CcTet D337F mutant protein retained the catalytic activity on 6mA but lost activity on 5-methylcytosine. Our findings uncovered a 6mA demethylase that works on dsDNA, suggesting potential 6mA demethylation in fungi and elucidating 6mA recognition and the catalytic mechanism of CcTet. The CcTet D337F mutant protein also provides a chemical biology tool for future functional manipulation of DNA 6mA in vivo.