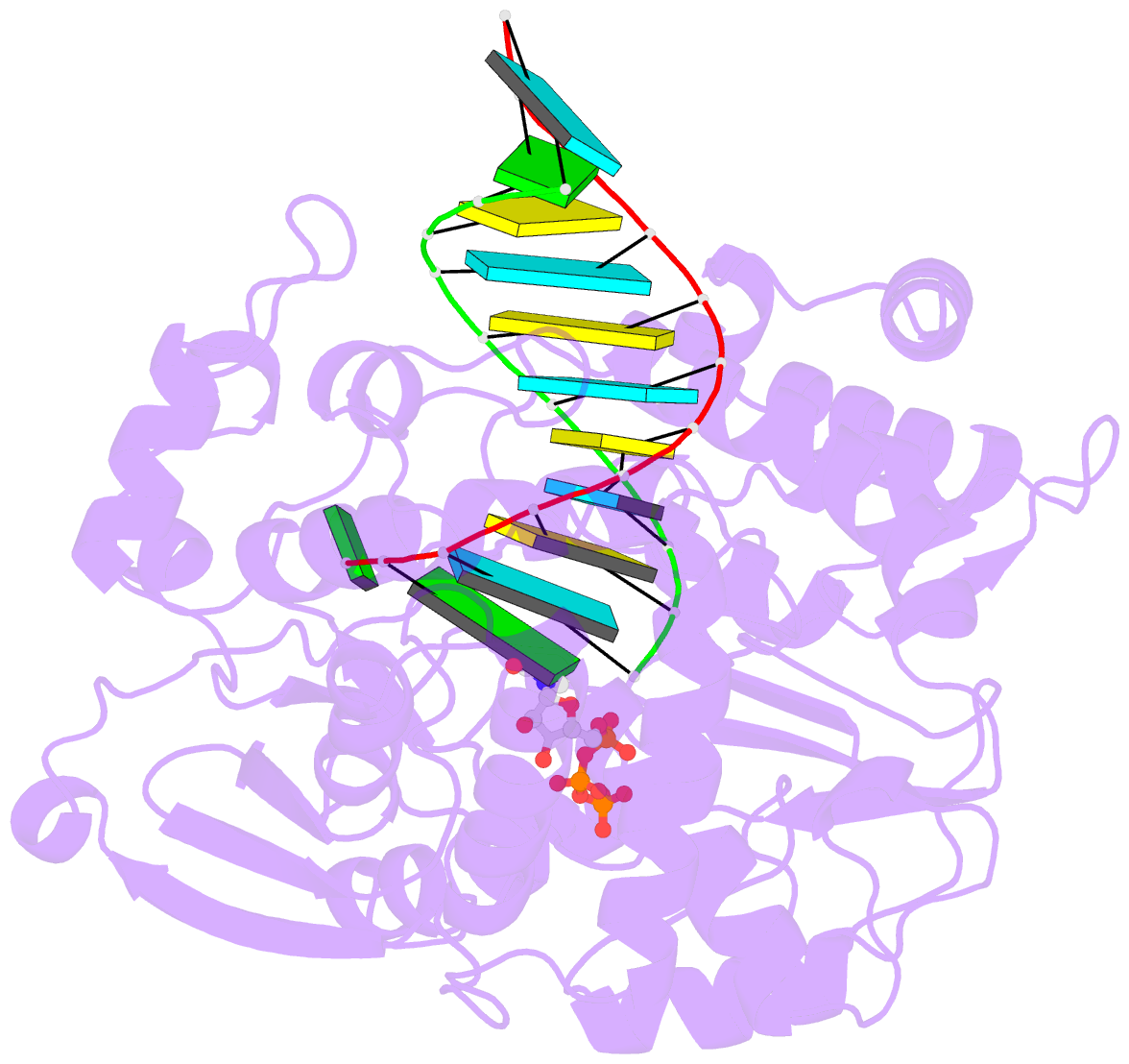
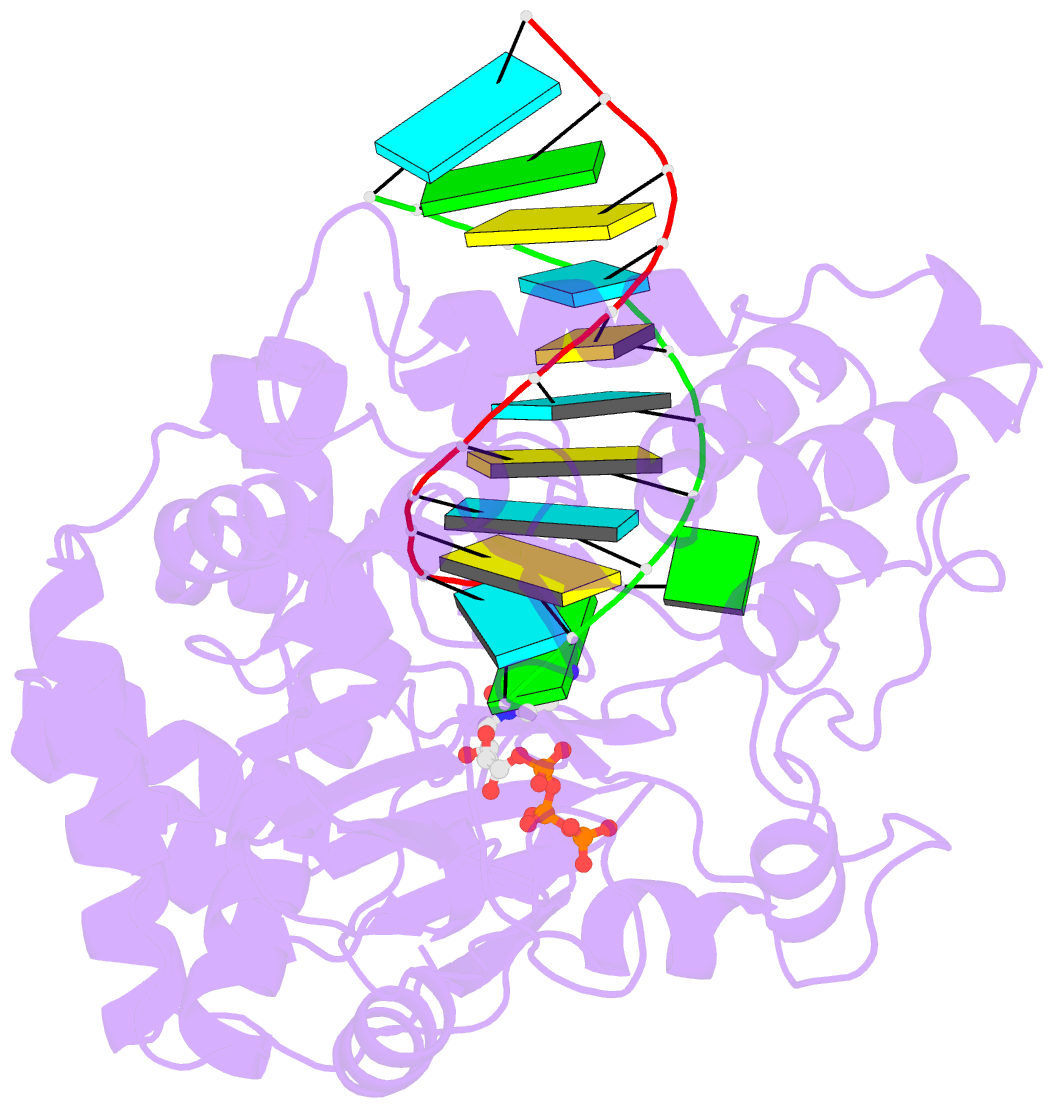
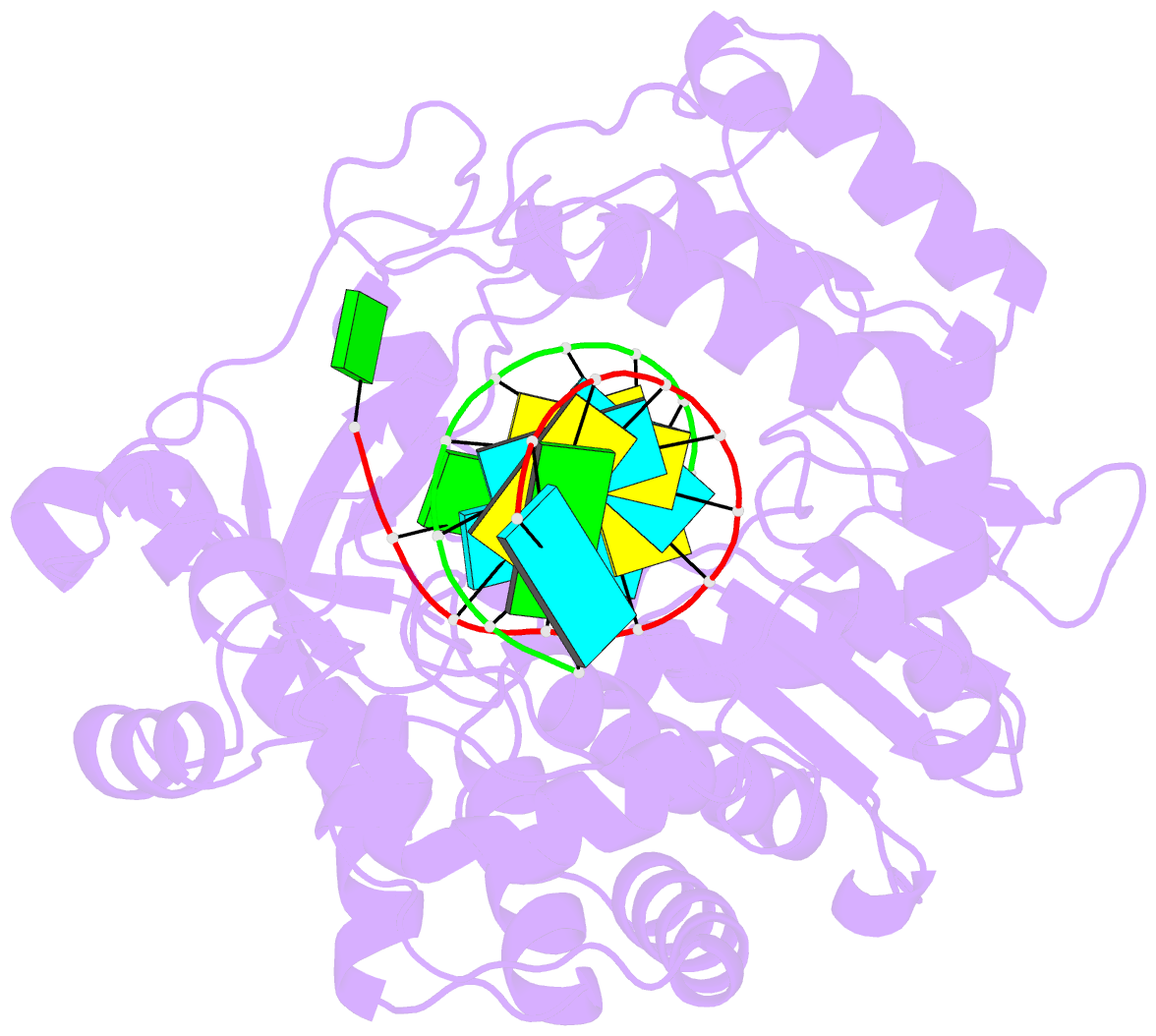
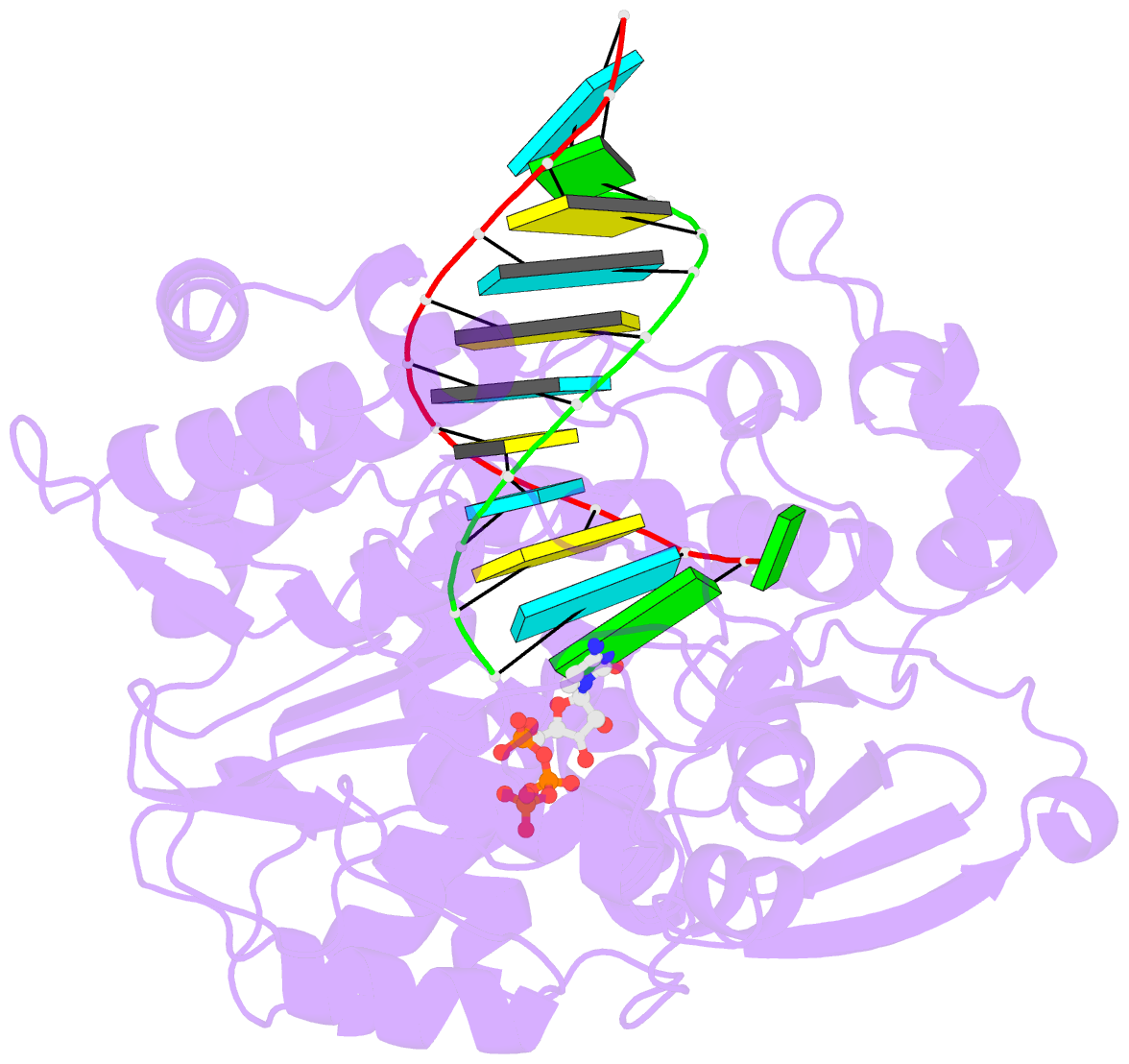
Summary information and primary citation
- PDB-id
- 7w9s; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transferase-RNA
- Method
- X-ray (2.53 Å)
- Summary
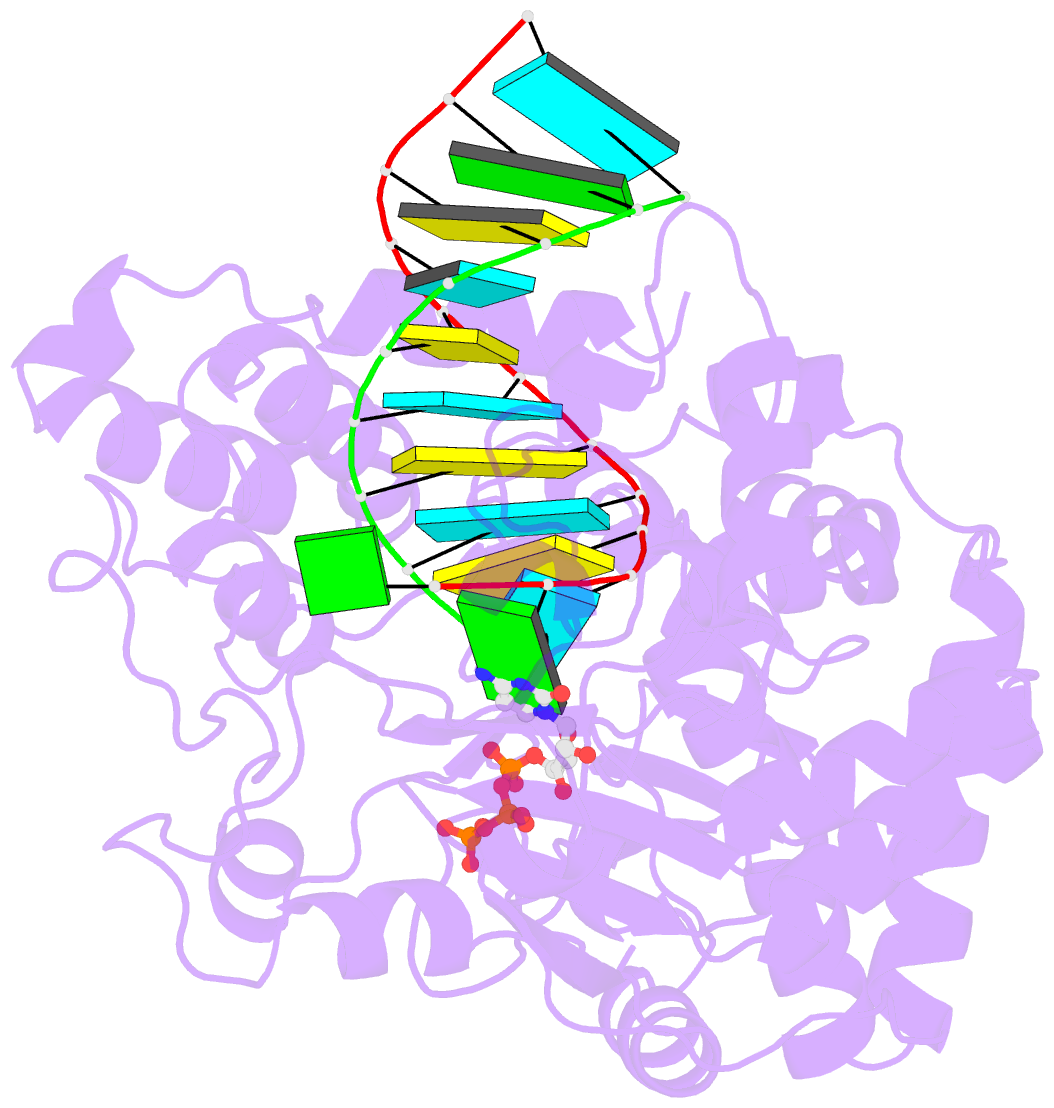
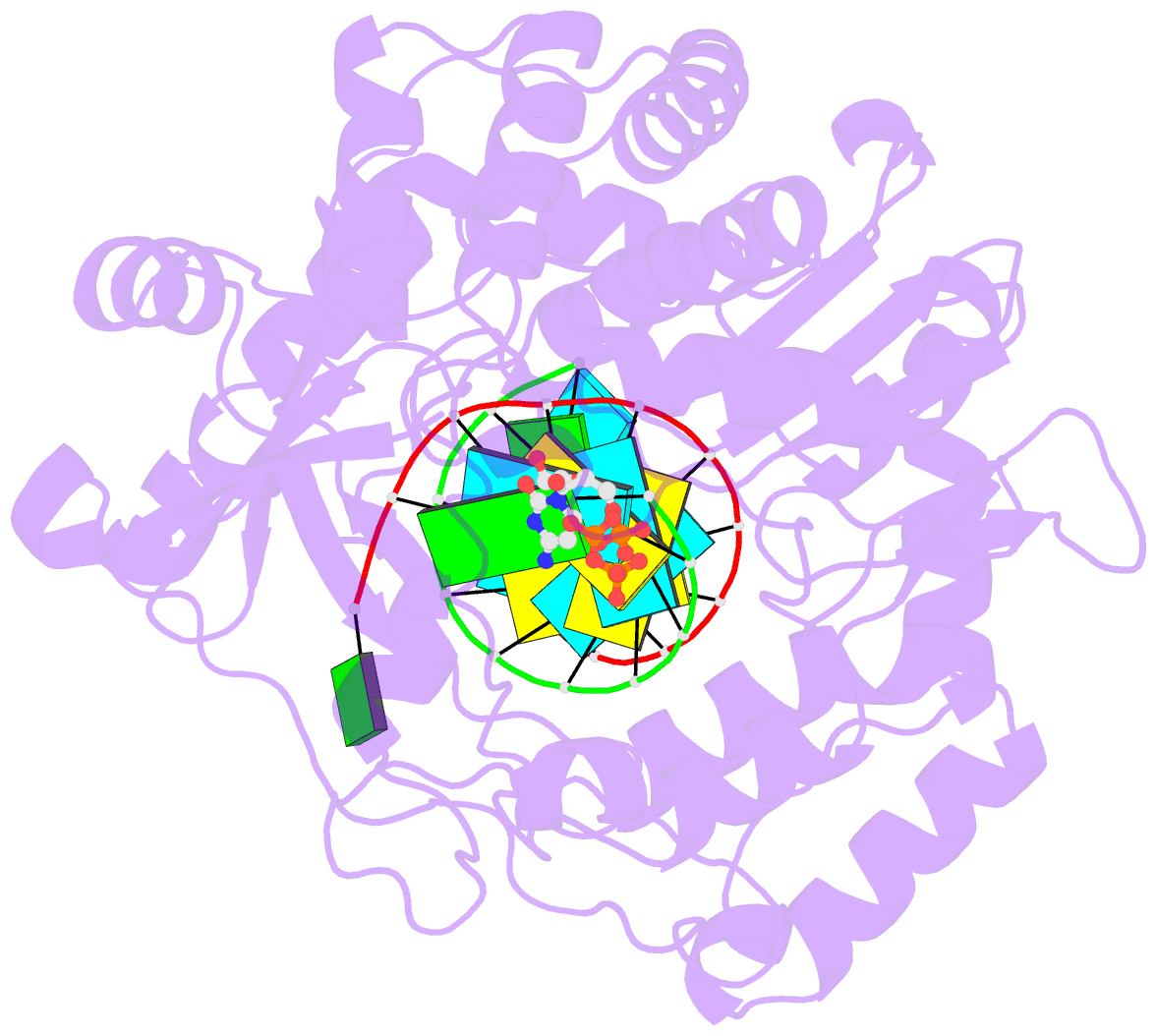
- Crystal structure of the enterovirus 71 polymerase elongation complex (c1s3 form)
- Reference
- Li R, Wang M, Gong P (2022): "Crystal structure of a pre-chemistry viral RNA-dependent RNA polymerase suggests participation of two basic residues in catalysis." Nucleic Acids Res., 50, 12389-12399. doi: 10.1093/nar/gkac1133.
- Abstract
- The nucleic acid polymerase-catalyzed nucleotidyl transfer reaction associated with polymerase active site closure is a key step in the nucleotide addition cycle (NAC). Two proton transfer events can occur in such a nucleotidyl transfer: deprotonation of the priming nucleotide 3'-hydroxyl nucleophile and protonation of the pyrophosphate (PPi) leaving group. In viral RNA-dependent RNA polymerases (RdRPs), whether and how active site residues participate in this two-proton transfer reaction remained to be clarified. Here we report a 2.5 Å resolution crystal structure of enterovirus 71 (EV71) RdRP in a catalytically closed pre-chemistry conformation, with a proposed proton donor candidate K360 in close contact with the NTP γ-phosphate. Enzymology data reveal that K360 mutations not only reduce RdRP catalytic efficiency but also alter pH dependency profiles in both elongation and pre-elongation synthesis modes. Interestingly, mutations at R174, an RdRP-invariant residue in motif F, had similar effects with additional impact on the Michaelis constant of NTP (KM,NTP). However, direct participation in protonation was not evident for K360 or R174. Our data suggest that both K360 and R174 participate in nucleotidyl transfer, while their possible roles in acid-base or positional catalysis are discussed in comparison with other classes of nucleic acid polymerases.