Summary information and primary citation
- PDB-id
- 7w9v; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- gene regulation
- Method
- cryo-EM (3.95 Å)
- Summary
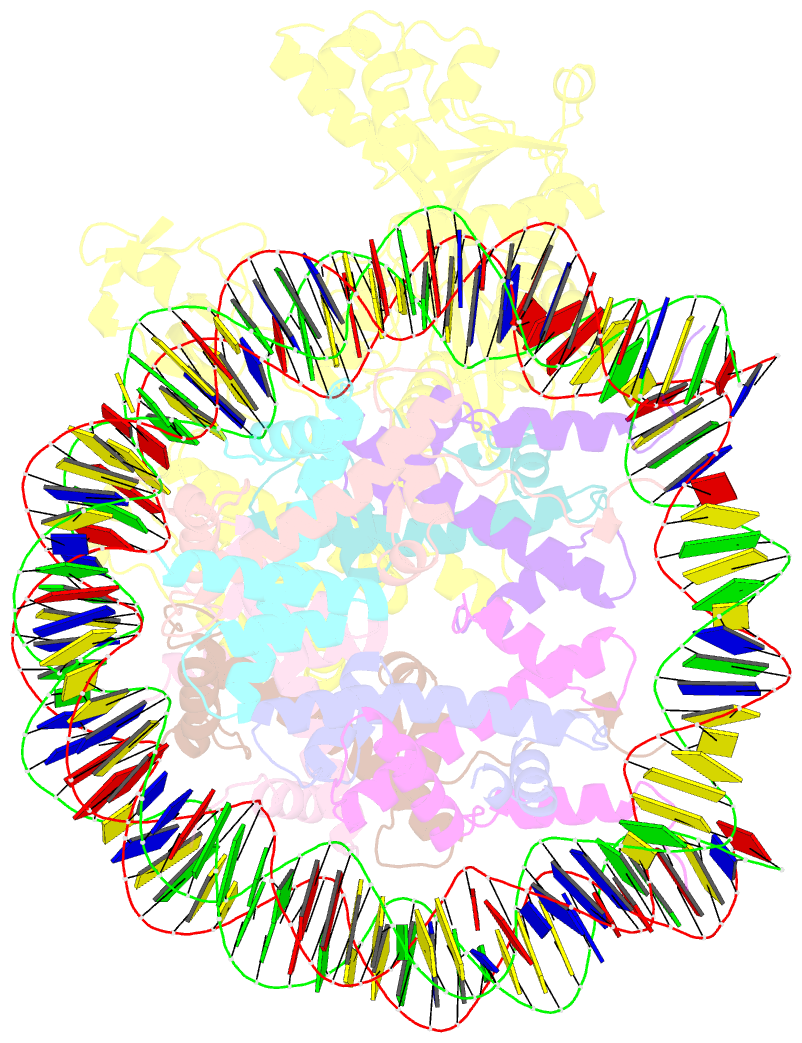
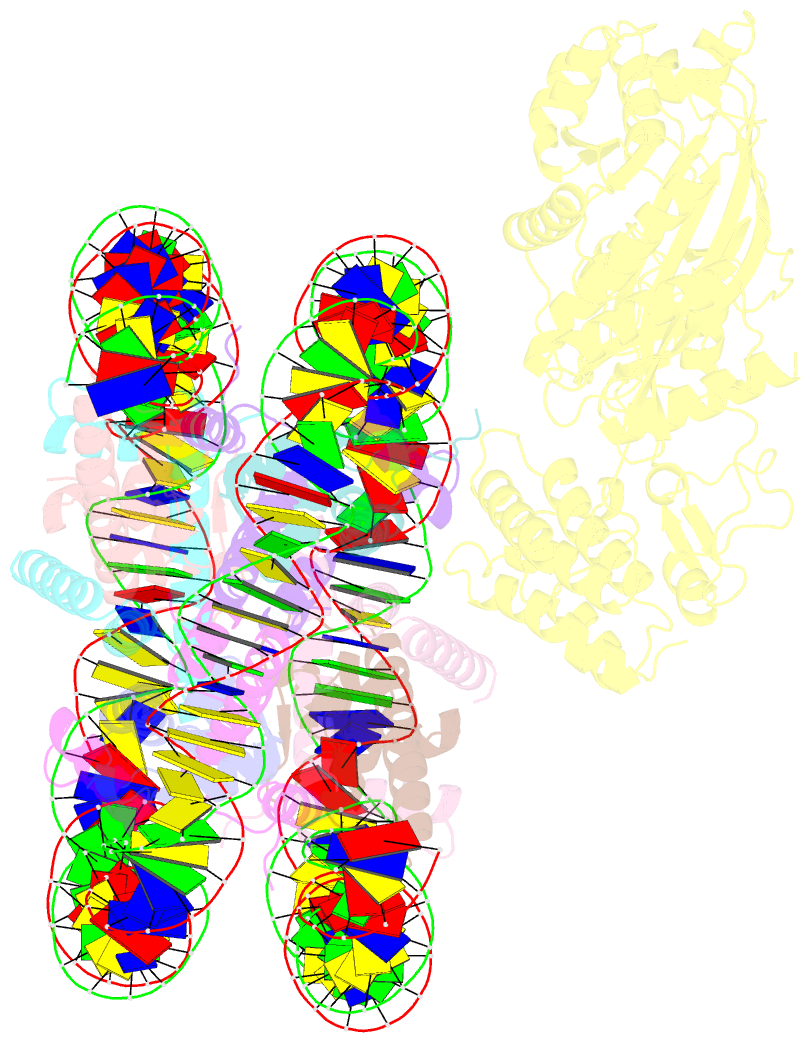
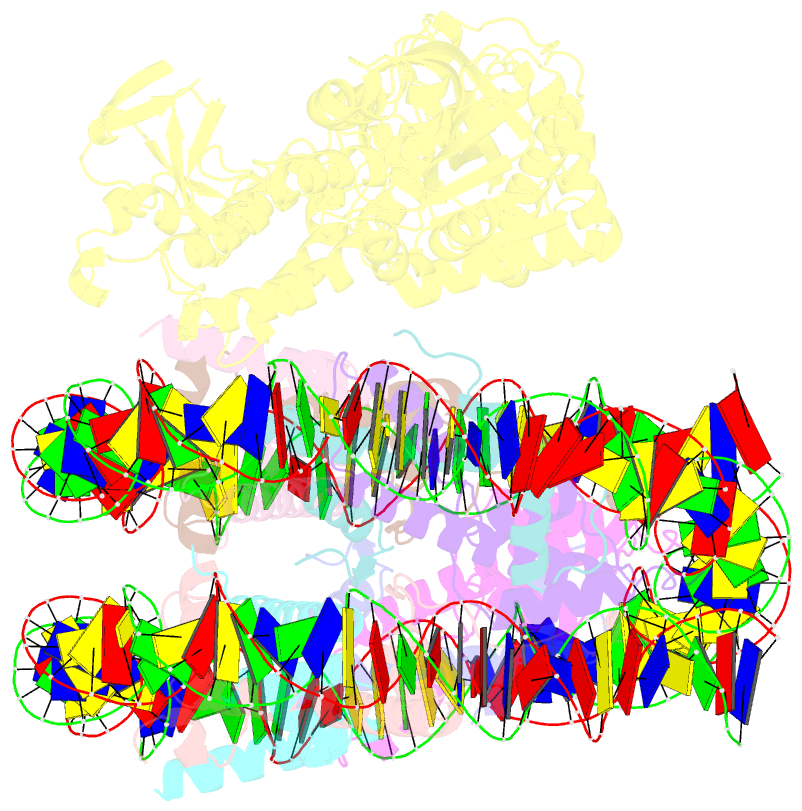
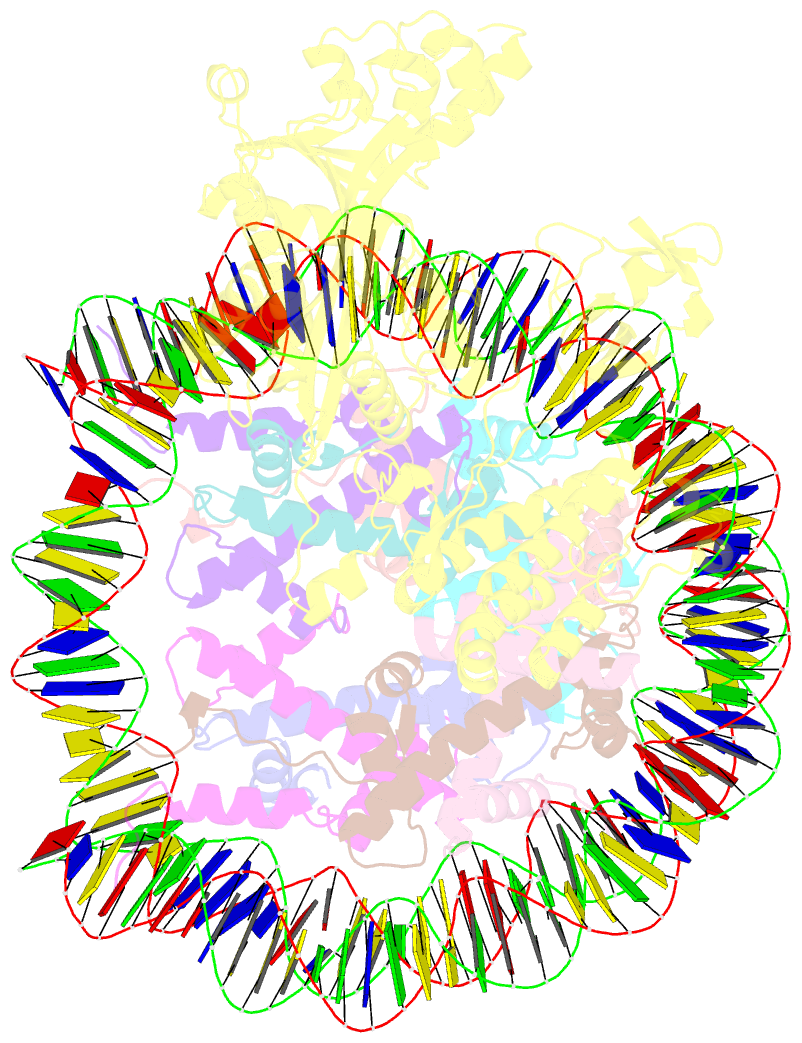
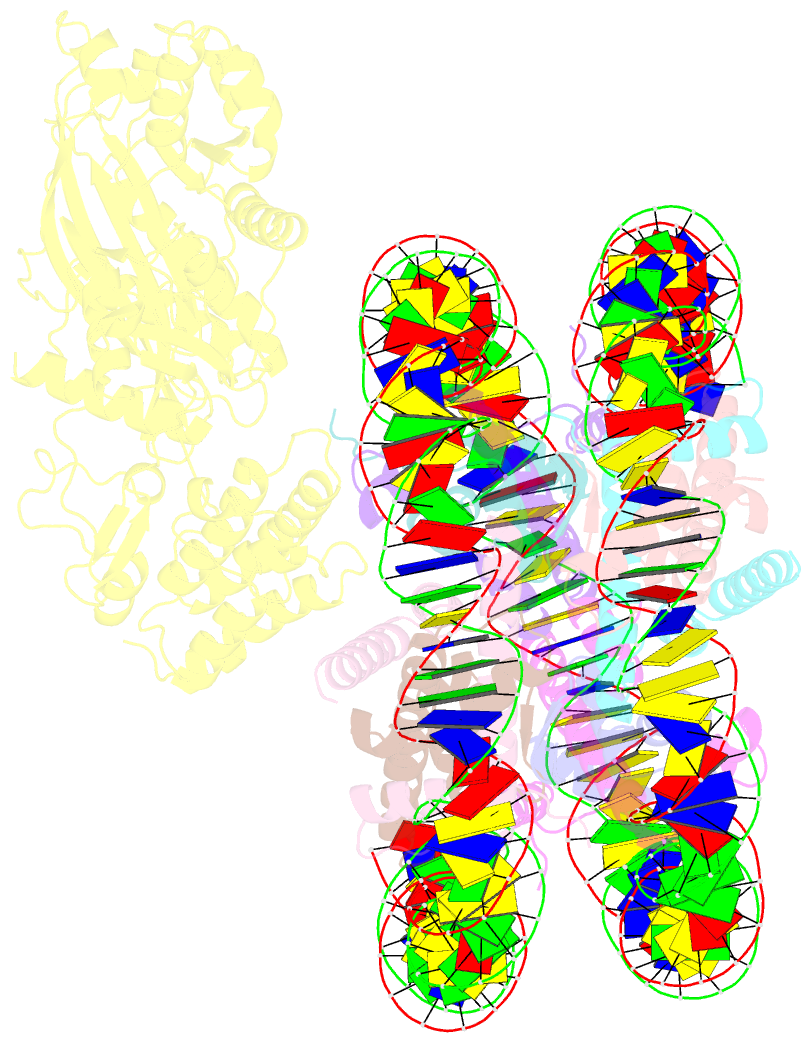
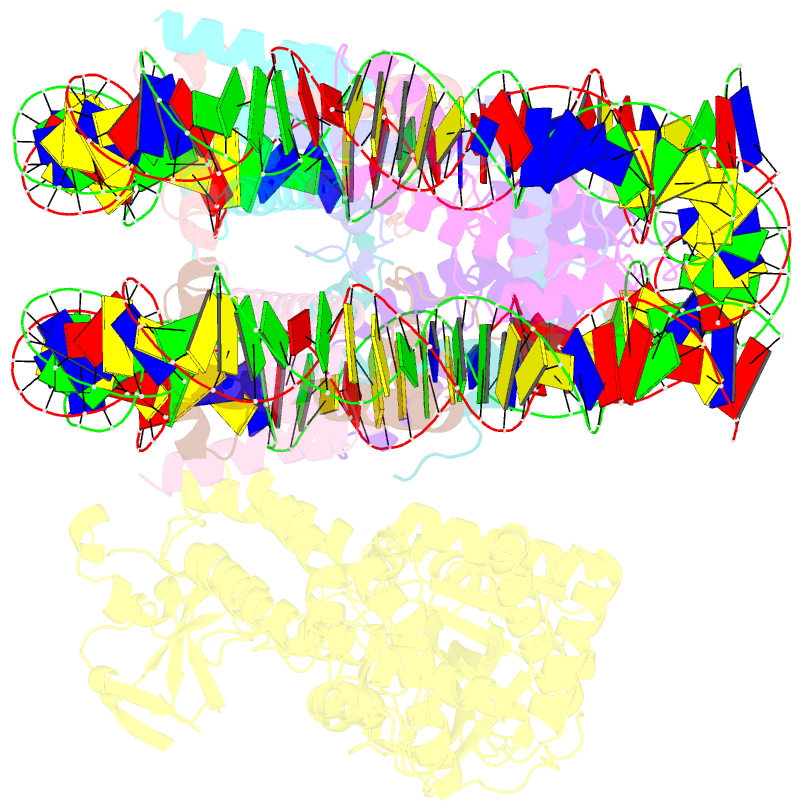
- cryo-EM structure of nucleosome in complex with p300 acetyltransferase catalytic core (complex i)
- Reference
- Hatazawa S, Liu J, Takizawa Y, Zandian M, Negishi L, Kutateladze TG, Kurumizaka H (2022): "Structural basis for binding diversity of acetyltransferase p300 to the nucleosome." Iscience, 25, 104563. doi: 10.1016/j.isci.2022.104563.
- Abstract
- p300 is a human acetyltransferase that associates with chromatin and mediates vital cellular processes. We now report the cryo-electron microscopy structures of the p300 catalytic core in complex with the nucleosome core particle (NCP). In the most resolved structure, the HAT domain and bromodomain of p300 contact nucleosomal DNA at superhelical locations 2 and 3, and the catalytic site of the HAT domain are positioned near the N-terminal tail of histone H4. Mutations of the p300-DNA interfacial residues of p300 substantially decrease binding to NCP. Three additional classes of p300-NCP complexes show different modes of the p300-NCP complex formation. Our data provide structural details critical to our understanding of the mechanism by which p300 acetylates multiple sites on the nucleosome.