Summary information and primary citation
- PDB-id
- 7wb1; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- RNA binding protein-RNA-DNA
- Method
- cryo-EM (3.7 Å)
- Summary
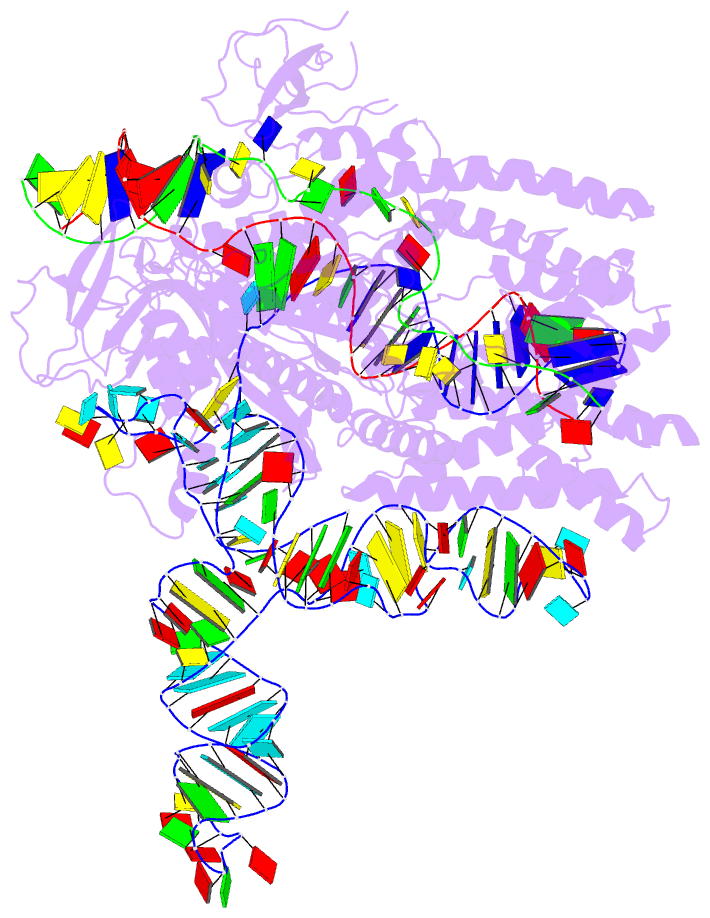
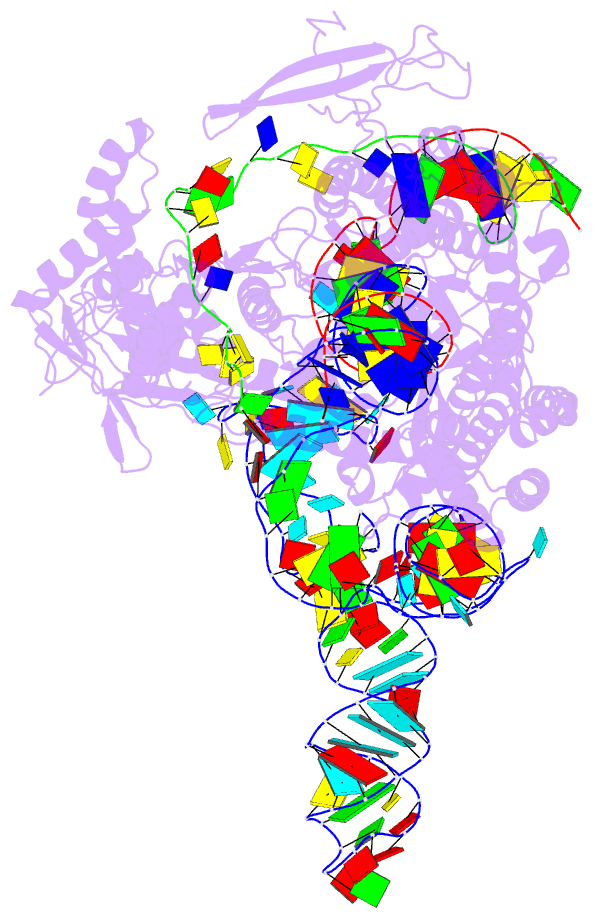
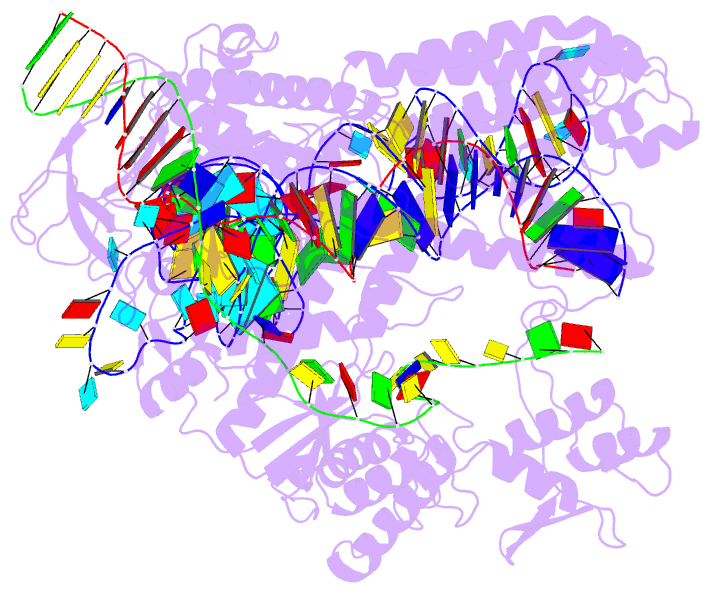
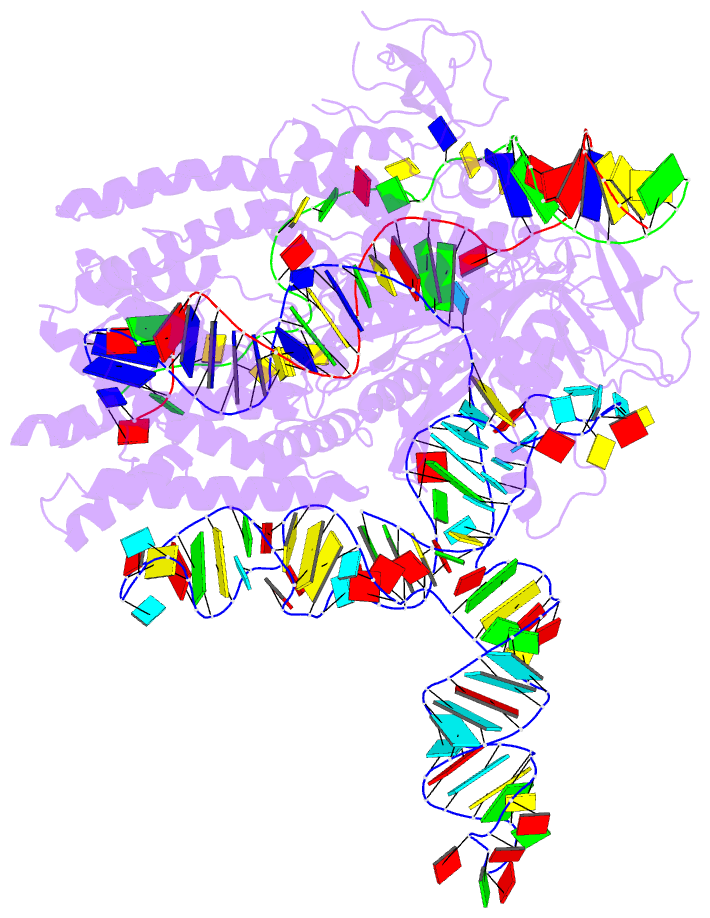
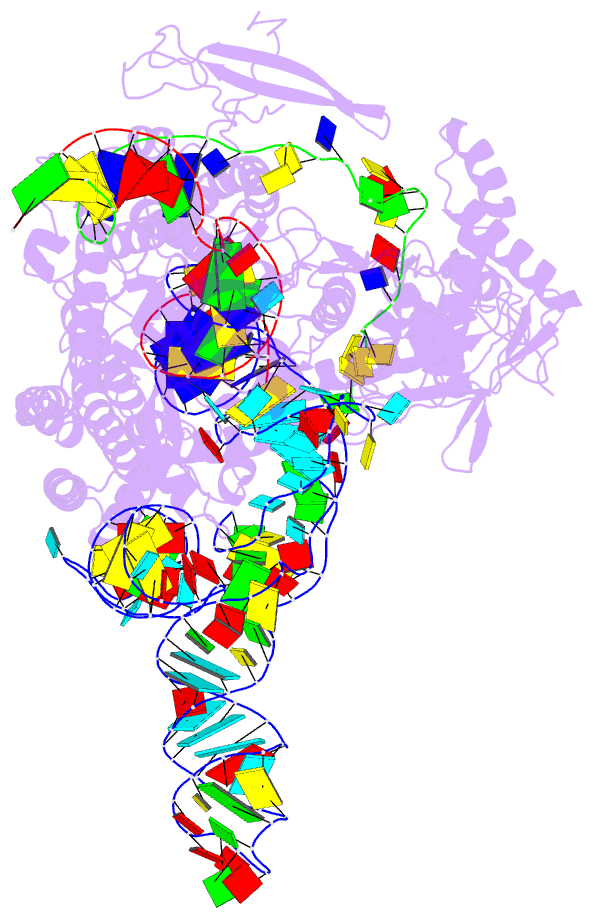
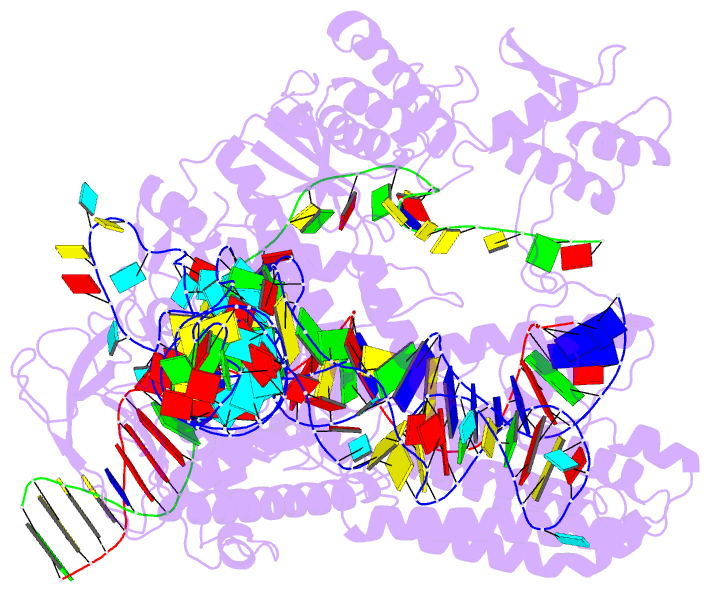
- Plmcasx-sgrnav2-dsDNA ternary complex at nts loading state
- Reference
- Tsuchida CA, Zhang S, Doost MS, Zhao Y, Wang J, O'Brien E, Fang H, Li CP, Li D, Hai ZY, Chuck J, Brotzmann J, Vartoumian A, Burstein D, Chen XW, Nogales E, Doudna JA, Liu JG (2022): "Chimeric CRISPR-CasX enzymes and guide RNAs for improved genome editing activity." Mol.Cell, 82, 1199-1209.e6. doi: 10.1016/j.molcel.2022.02.002.
- Abstract
- A compact protein with a size of <1,000 amino acids, the CRISPR-associated protein CasX is a fundamentally distinct RNA-guided nuclease when compared to Cas9 and Cas12a. Although it can induce RNA-guided genome editing in mammalian cells, the activity of CasX is less robust than that of the widely used S. pyogenes Cas9. Here, we show that structural features of two CasX homologs and their guide RNAs affect the R-loop complex assembly and DNA cleavage activity. Cryo-EM-based structural engineering of either the CasX protein or the guide RNA produced two new CasX genome editors (DpbCasX-R3-v2 and PlmCasX-R1-v2) with significantly improved DNA manipulation efficacy. These results advance both the mechanistic understanding of CasX and its application as a genome-editing tool.