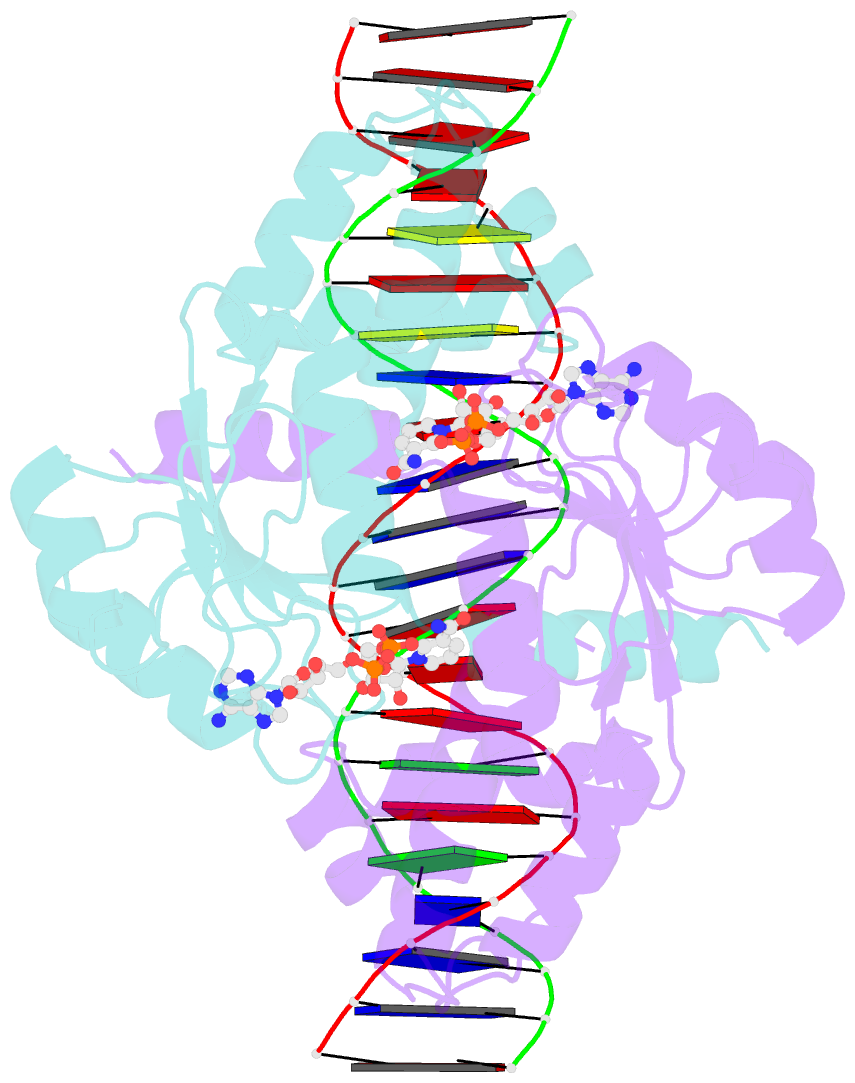
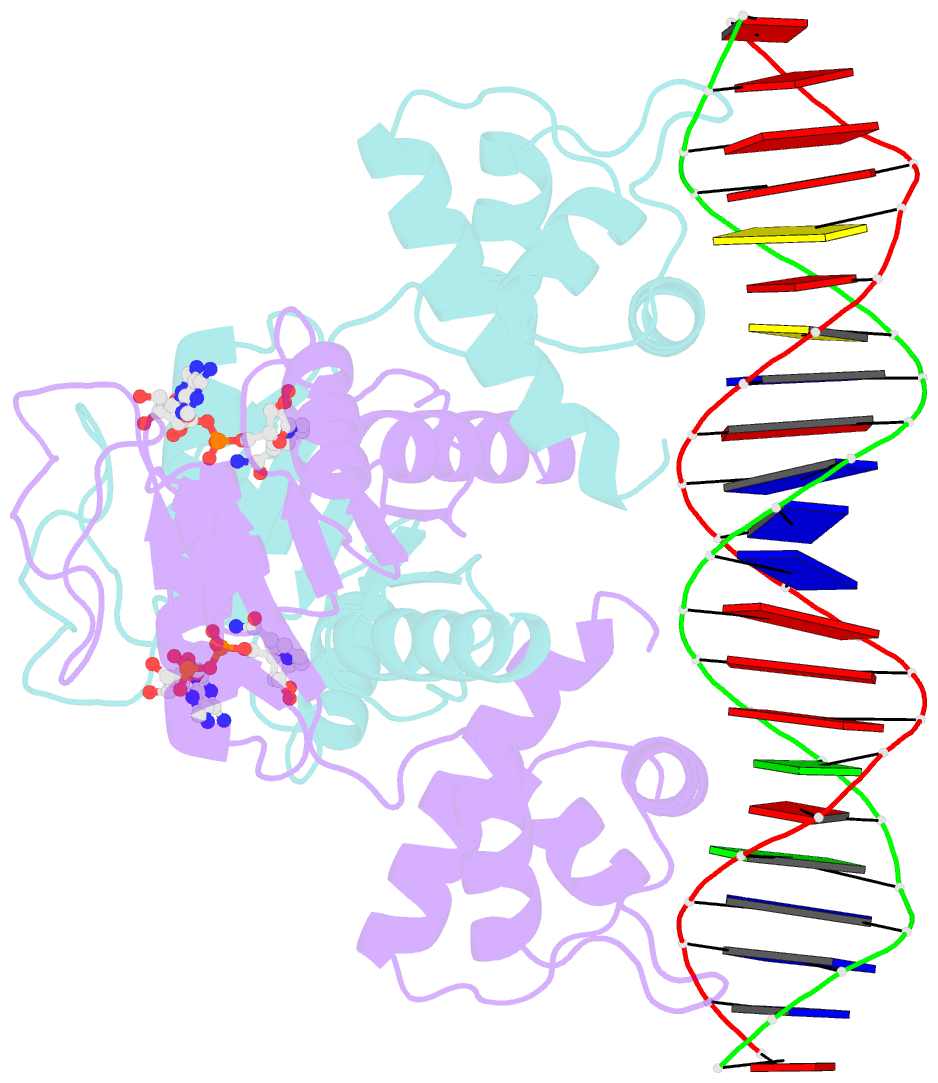
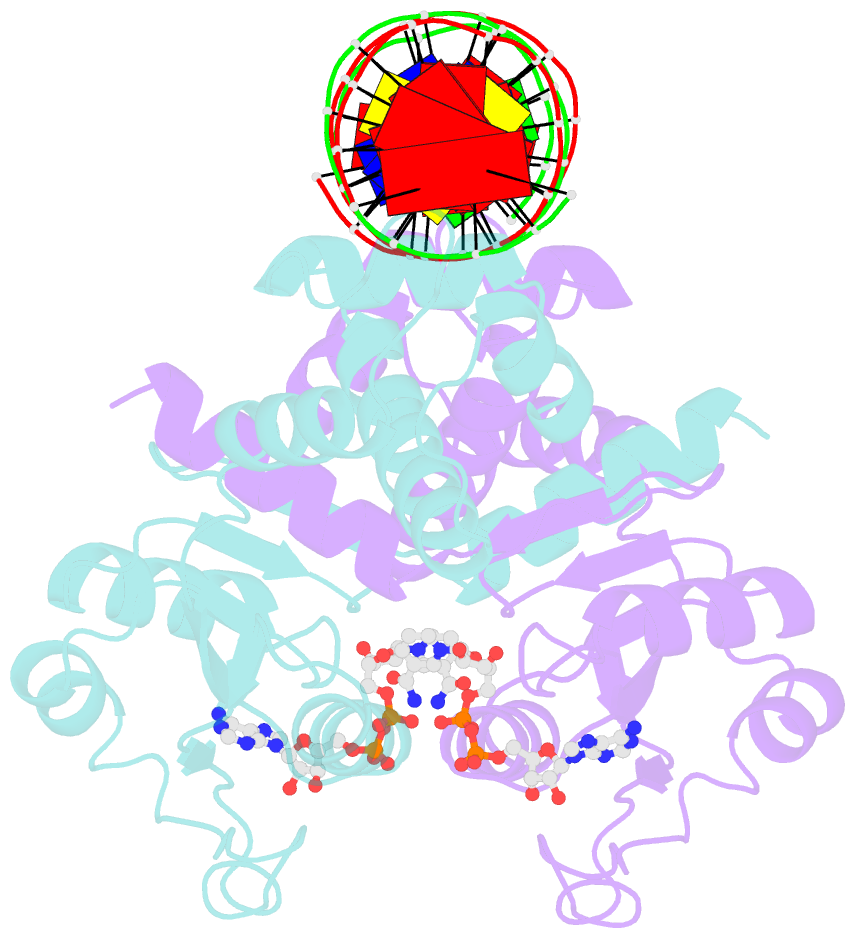
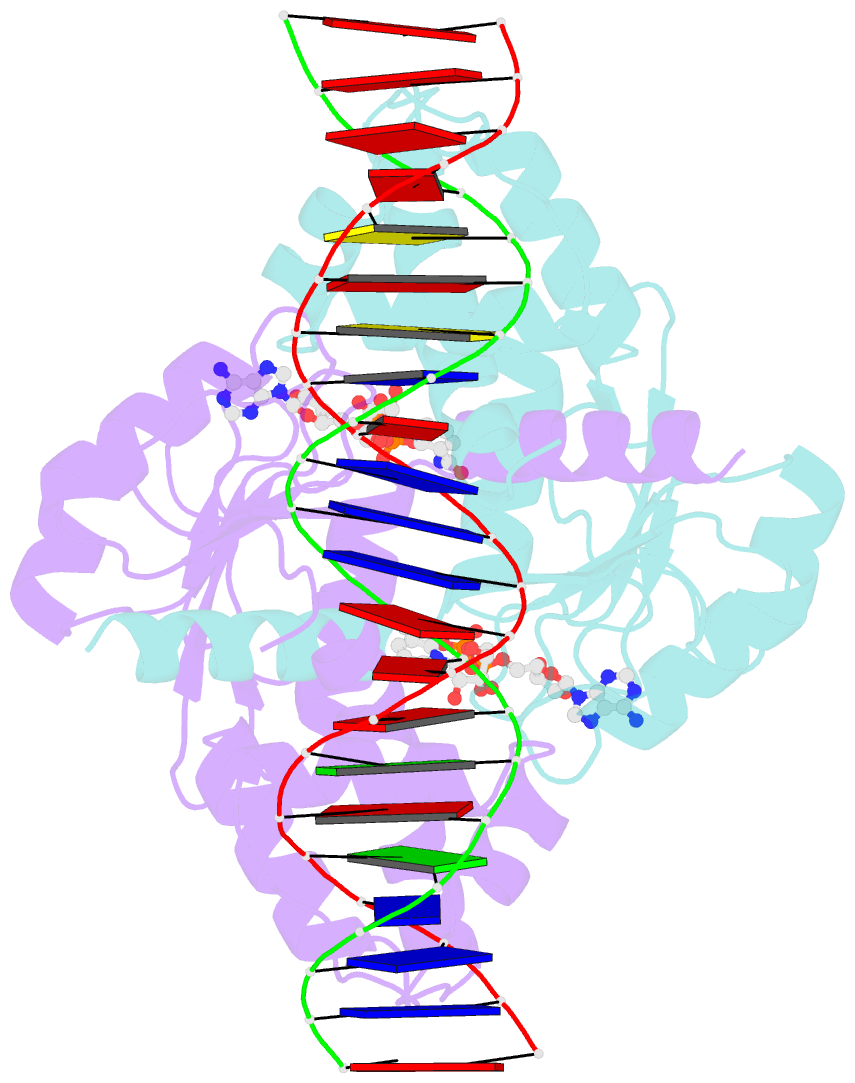
Summary information and primary citation
- PDB-id
- 7wb3; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- gene regulation-DNA
- Method
- X-ray (2.401 Å)
- Summary
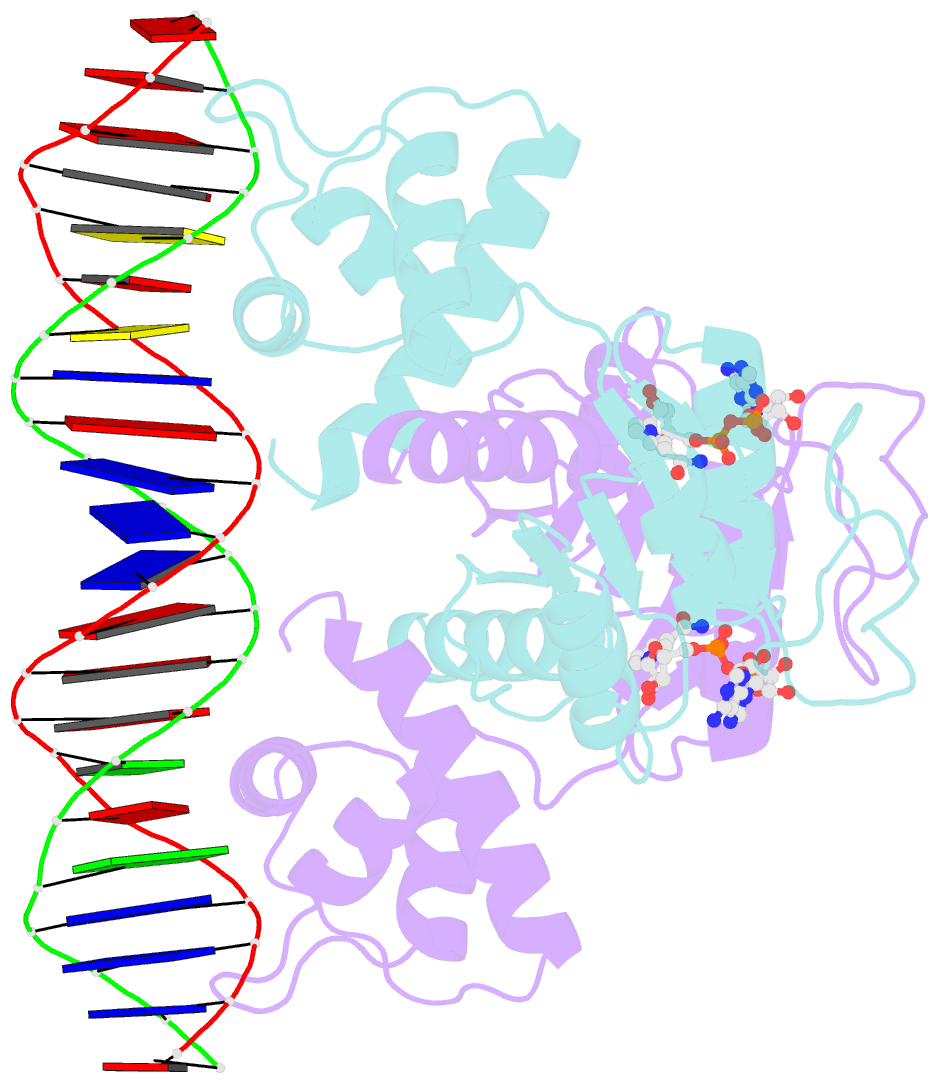
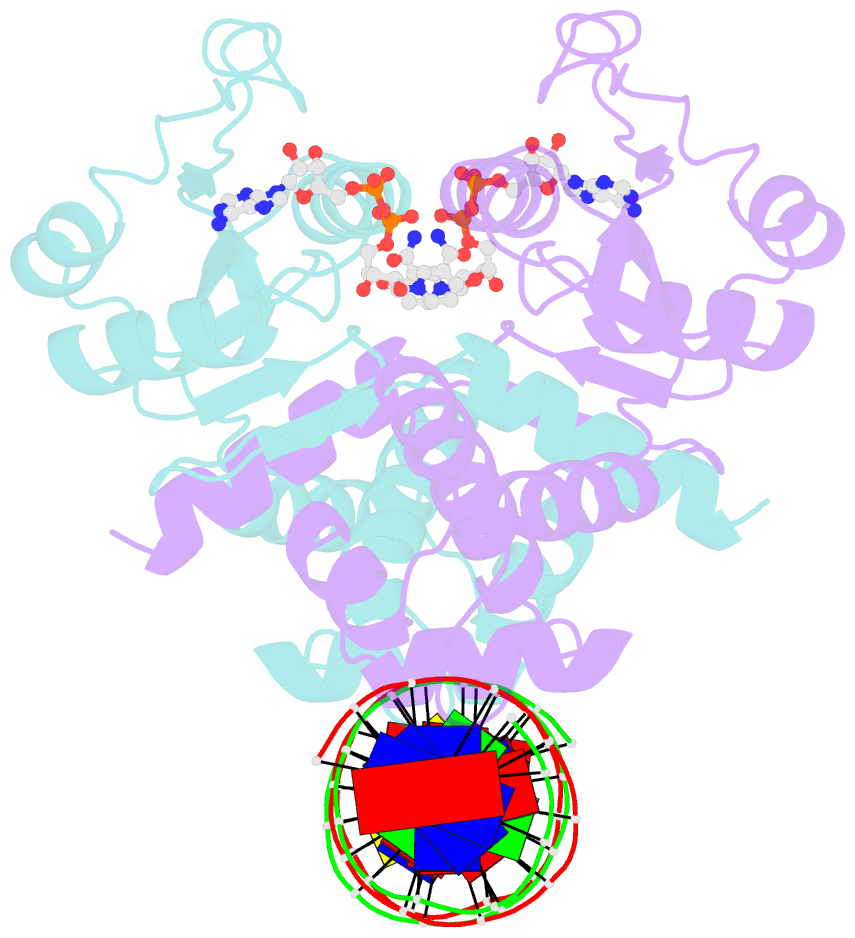
- Crystal structure of t. maritima rex in ternary complex
- Reference
- Jeong KH, Lee HJ, Park YW, Lee JY (2022): "Structural Basis of Redox-Sensing Transcriptional Repressor Rex with Cofactor NAD + and Operator DNA." Int J Mol Sci, 23. doi: 10.3390/ijms23031578.
- Abstract
- The transcriptional repressor Rex plays important roles in regulating the expression of respiratory genes by sensing the reduction-oxidation (redox) state according to the intracellular NAD+/NADH balance. Previously, we reported on crystal structures of apo, NAD+-bound, and NADH-bound forms of Rex from Thermotoga maritima to analyze the structural basis of transcriptional regulation depending on either NAD+ or NADH binding. In this study, the crystal structure of Rex in ternary complex with NAD+ and operator DNA revealed that the N-terminal domain of Rex, including the helix-turn-helix motif, forms extensive contacts with DNA in addition to DNA sequence specificity. Structural comparison of the Rex in apo, NAD+-bound, NADH-bound, and ternary complex forms provides a comprehensive picture of transcriptional regulation in the Rex. These data demonstrate that the conformational change in Rex when binding with the reduced NADH or oxidized NAD+ determines operator DNA binding. The movement of the N-terminal domains toward the operator DNA was blocked upon binding of NADH ligand molecules. The structural results provide insights into the molecular mechanism of Rex binding with operator DNA and cofactor NAD+/NADH, which is conserved among Rex family repressors. Structural analysis of Rex from T. maritima also supports the previous hypothesis about the NAD+/NADH-specific transcriptional regulation mechanism of Rex homologues.