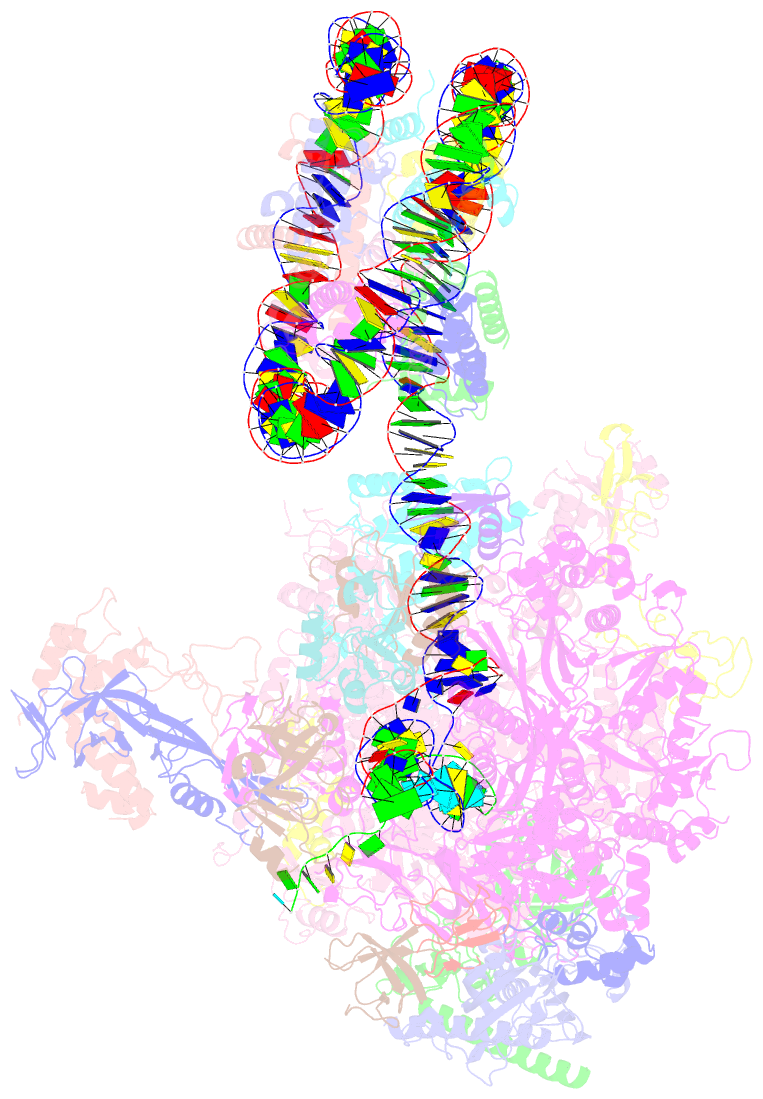
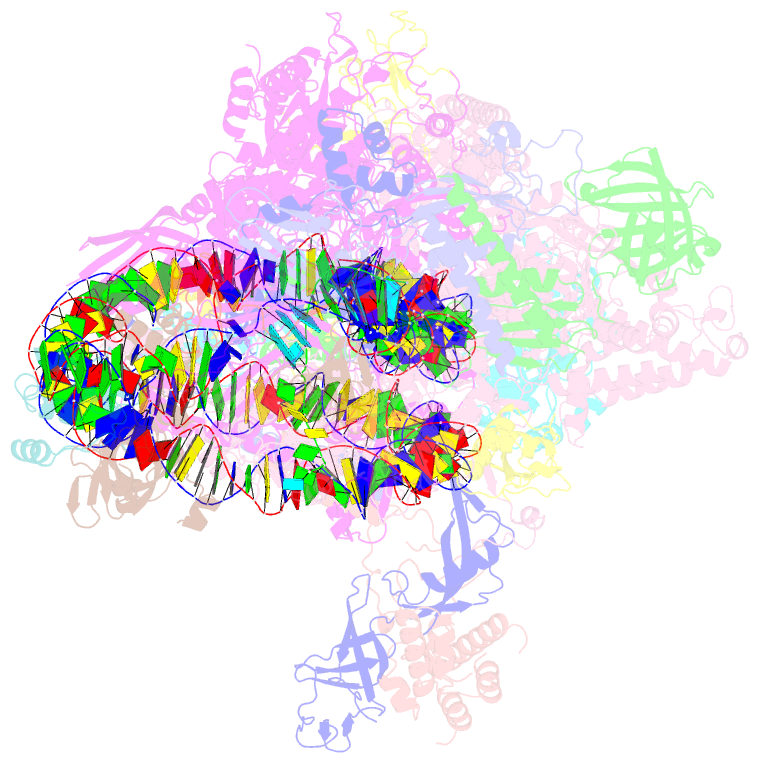
Summary information and primary citation
- PDB-id
- 7wbv; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription
- Method
- cryo-EM (4.1 Å)
- Summary
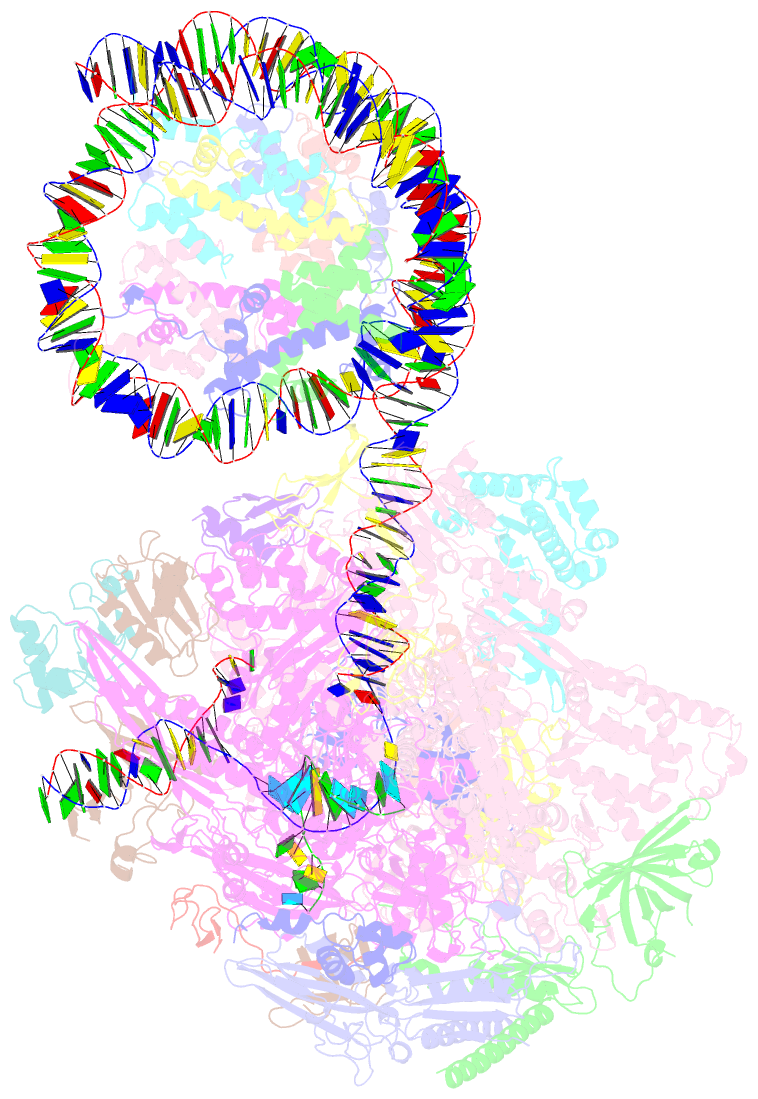
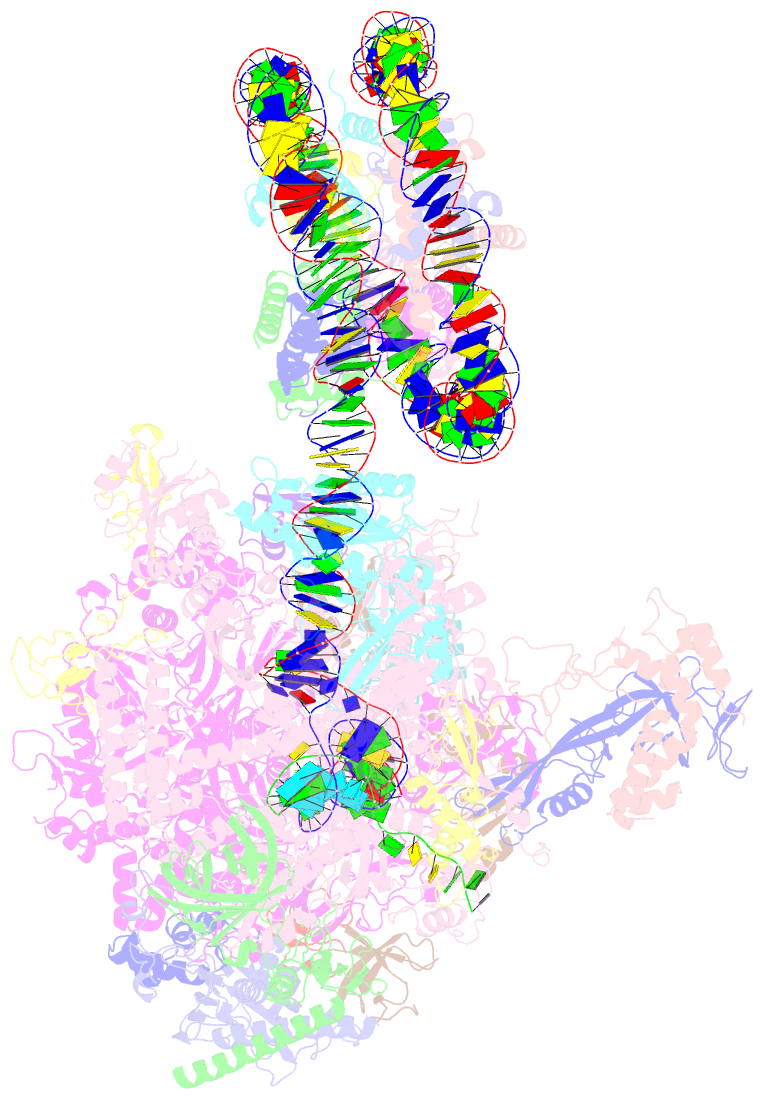
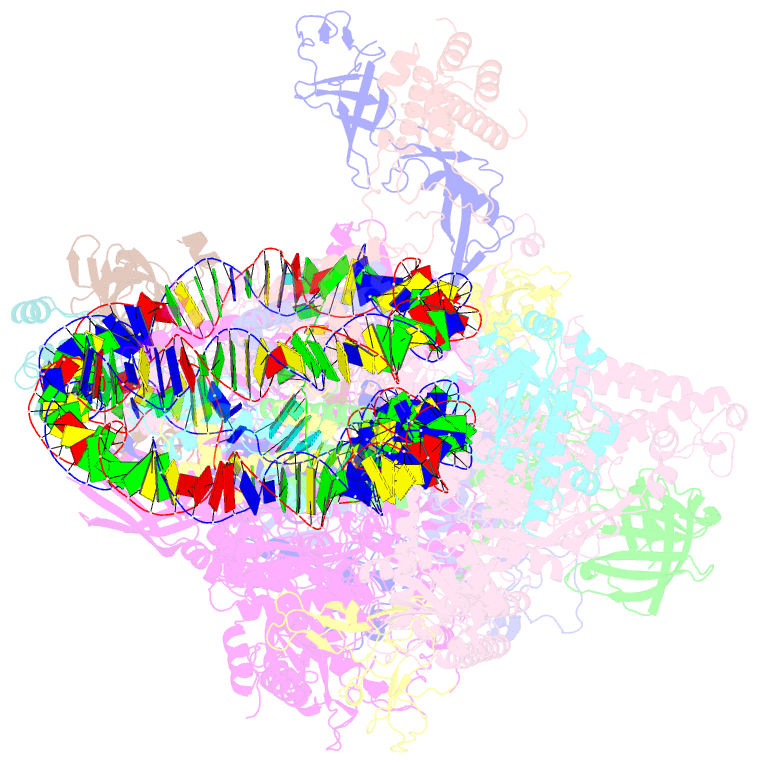
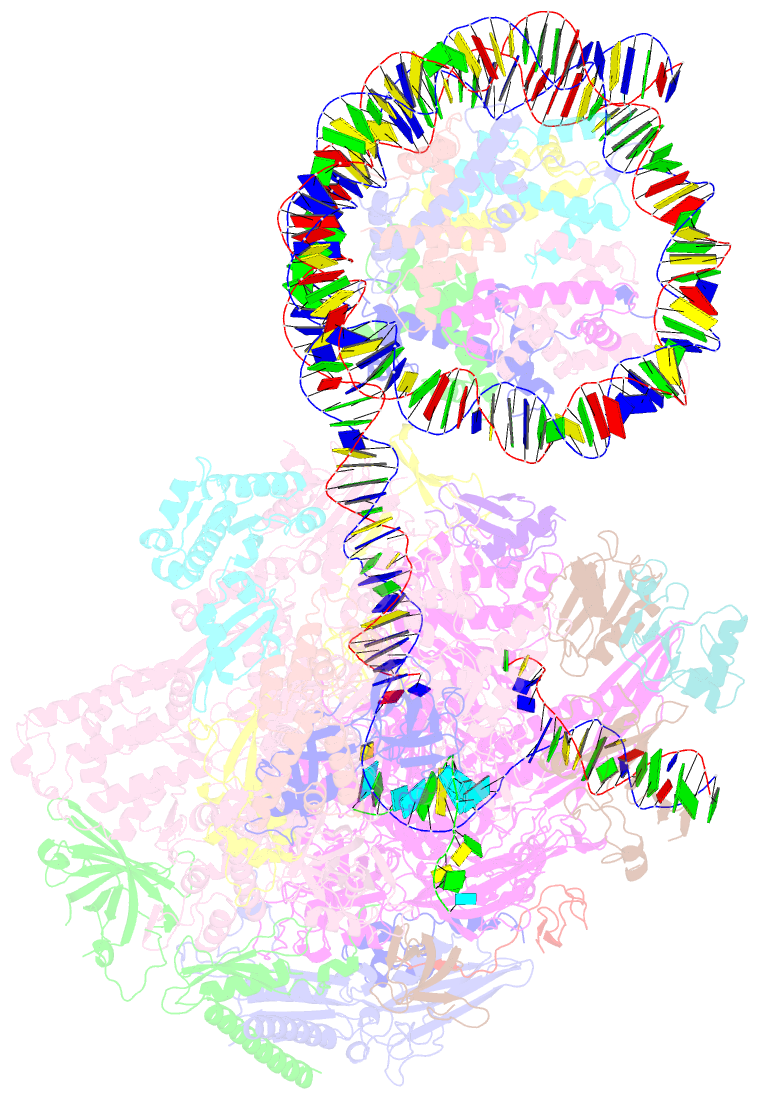
- RNA polymerase ii elongation complex bound with elf1 and spt4-5, stalled at shl(-4) of the nucleosome
- Reference
- Osumi K, Kujirai T, Ehara H, Ogasawara M, Kinoshita C, Saotome M, Kagawa W, Sekine SI, Takizawa Y, Kurumizaka H (2023): "Structural Basis of Damaged Nucleotide Recognition by Transcribing RNA Polymerase II in the Nucleosome." J.Mol.Biol., 435, 168130. doi: 10.1016/j.jmb.2023.168130.
- Abstract
- In transcription-coupled repair (TCR), transcribing RNA polymerase II (RNAPII) stalls at a DNA lesion and recruits TCR proteins to the damaged site. However, the mechanism by which RNAPII recognizes a DNA lesion in the nucleosome remains enigmatic. In the present study, we inserted an apurinic/apyrimidinic DNA lesion analogue, tetrahydrofuran (THF), in the nucleosomal DNA, where RNAPII stalls at the SHL(-4), SHL(-3.5), and SHL(-3) positions, and determined the structures of these complexes by cryo-electron microscopy. In the RNAPII-nucleosome complex stalled at SHL(-3.5), the nucleosome orientation relative to RNAPII is quite different from those in the SHL(-4) and SHL(-3) complexes, which have nucleosome orientations similar to naturally paused RNAPII-nucleosome complexes. Furthermore, we found that an essential TCR protein, Rad26 (CSB), enhances the RNAPII processivity, and consequently augments the DNA damage recognition efficiency of RNAPII in the nucleosome. The cryo-EM structure of the Rad26-RNAPII-nucleosome complex revealed that Rad26 binds to the stalled RNAPII through a novel interface, which is completely different from those previously reported. These structures may provide important information to understand the mechanism by which RNAPII recognizes the nucleosomal DNA lesion and recruits TCR proteins to the stalled RNAPII on the nucleosome.