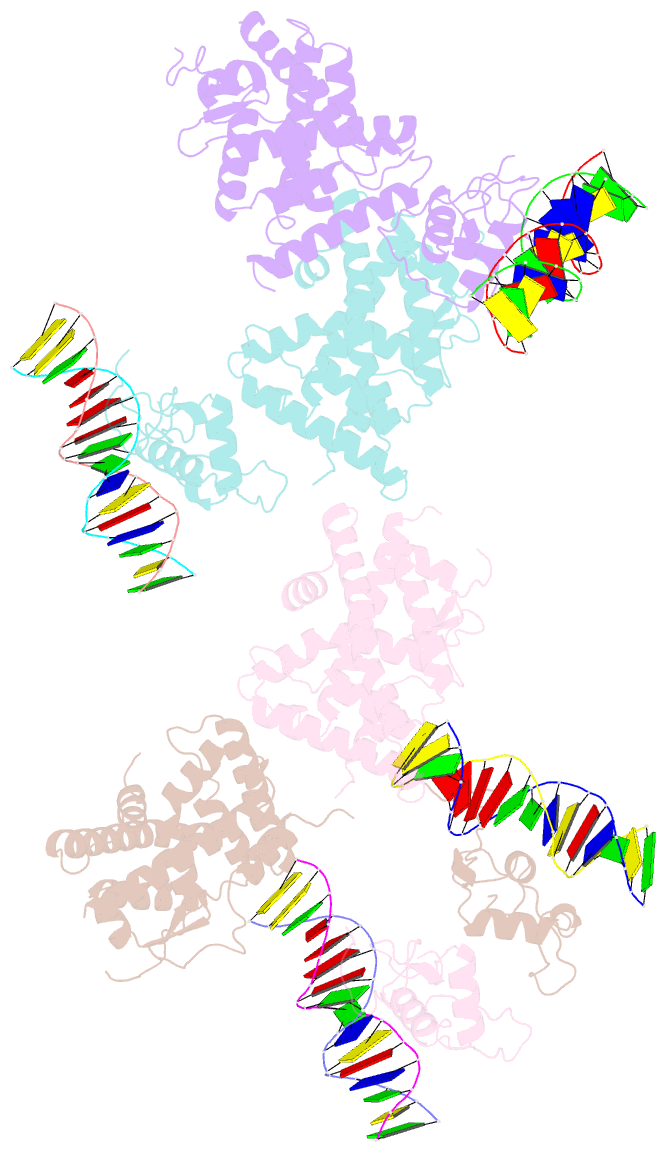
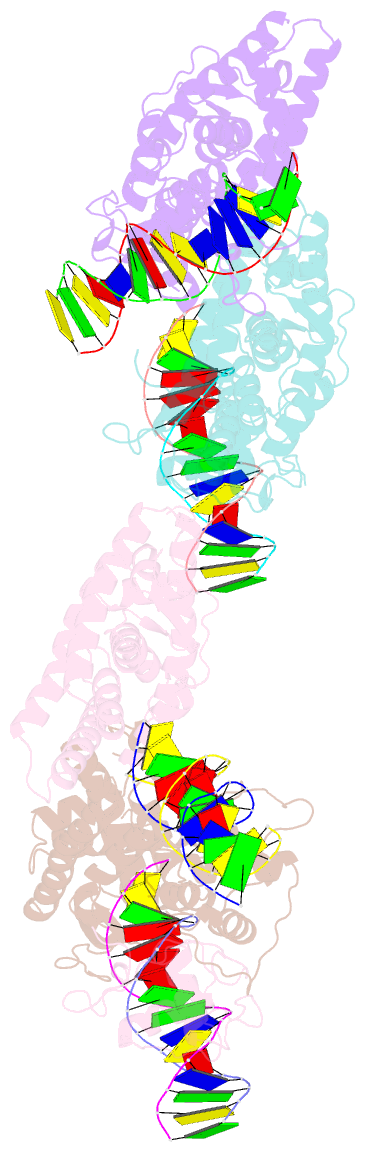
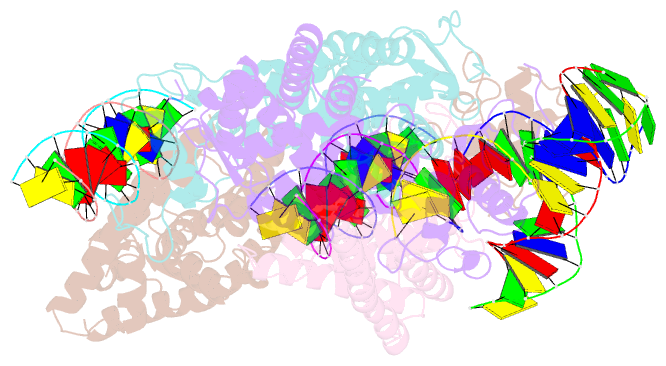
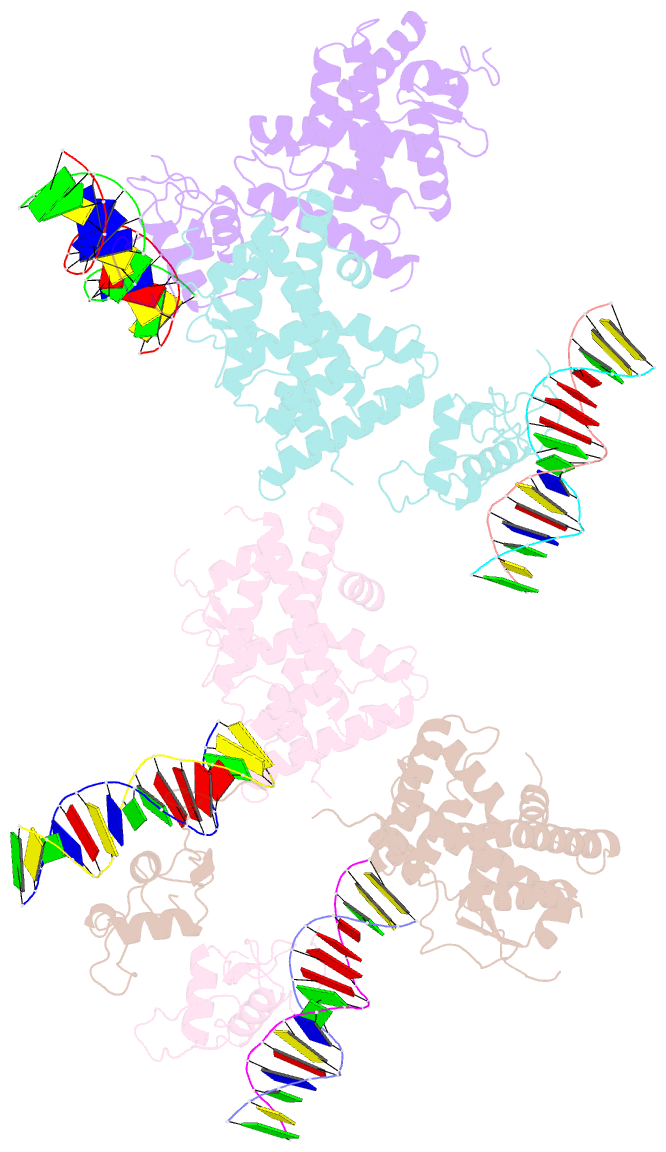
Summary information and primary citation
- PDB-id
- 7wnh; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription
- Method
- X-ray (3.1 Å)
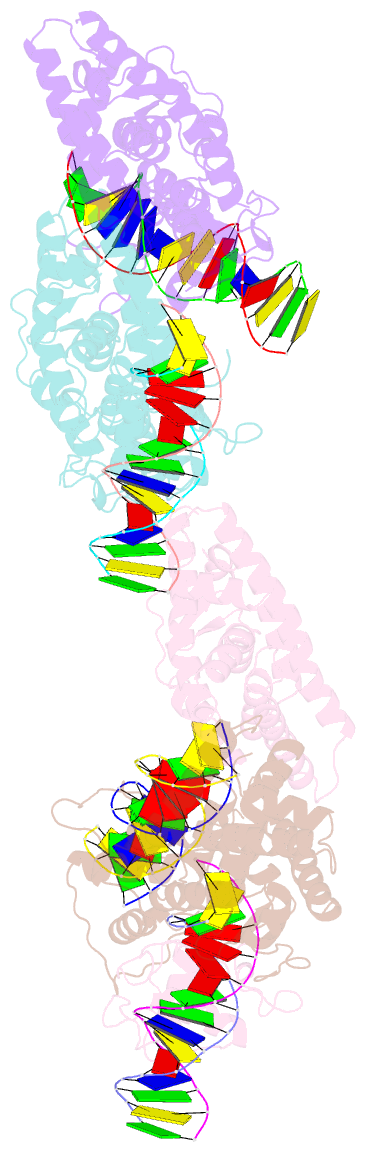
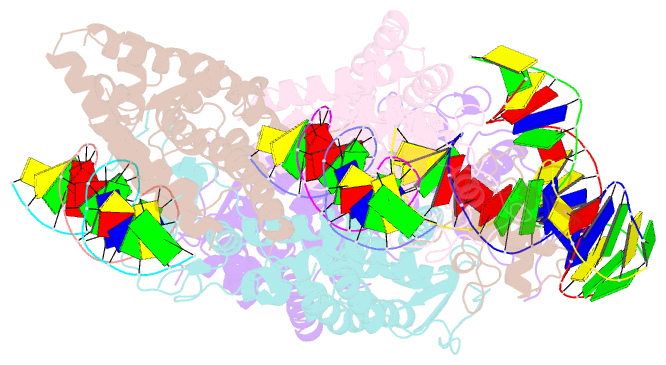
- Summary
- Crystal structure of nurr1 binding to nbre
- Reference
- Zhao M, Wang N, Guo Y, Li J, Yin Y, Dong Y, Zhu J, Peng C, Xu T, Liu J (2022): "Integrative analysis reveals structural basis for transcription activation of Nurr1 and Nurr1-RXR alpha heterodimer." Proc.Natl.Acad.Sci.USA, 119, e2206737119. doi: 10.1073/pnas.2206737119.
- Abstract
- Orphan nuclear receptor Nurr1 plays important roles in the progression of various diseases, including Parkinson's disease, neuroinflammation, Alzheimer's disease, and multiple sclerosis. It can recognize DNA as a monomer or heterodimer with retinoid X receptor α (RXRα). But the molecular mechanism of its transcriptional activity regulation is still largely unknown. Here we obtained a crystal structure of monomer Nurr1 (DNA- and ligand-binding domains, DBD and LBD) bound to NGFI-B response element. The structure exhibited two different forms with distinct DBD orientations, unveiling the conformational flexibility of nuclear receptor monomer. We then generated an integrative model of Nurr1-RXRα heterodimer. In the context of heterodimer, the structural flexibility of Nurr1 would contribute to its transcriptional activity modulation. We demonstrated that the DNA sequence may specifically modulate the transcriptional activity of Nurr1 in the absence of RXRα agonist, but the modulation can be superseded when the agonist binds to RXRα. Together, we propose a set of signaling pathways for the constitutive transcriptional activation of Nurr1 and provide molecular mechanisms for therapeutic discovery targeting Nurr1 and Nurr1-RXRα heterodimer.