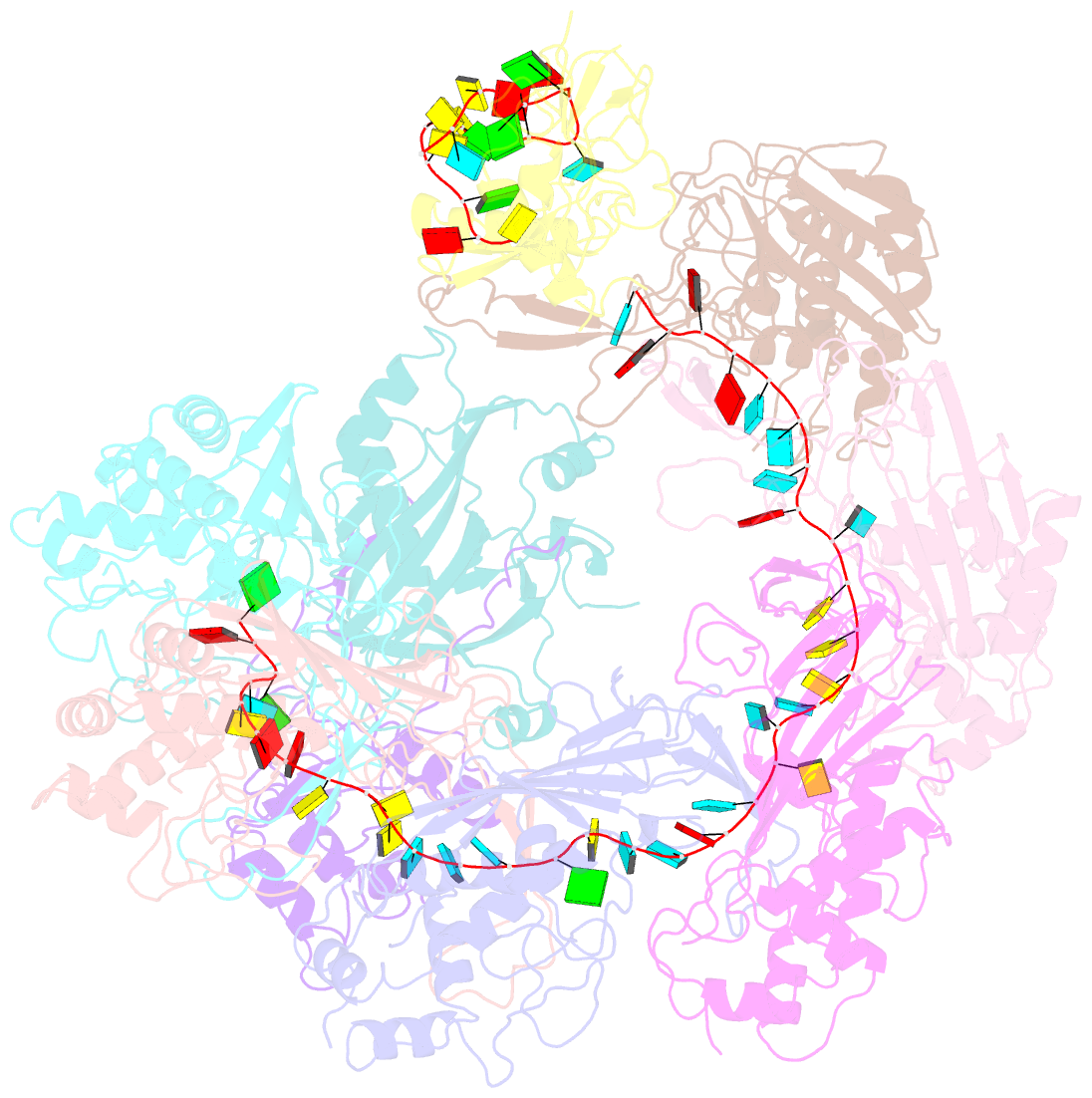
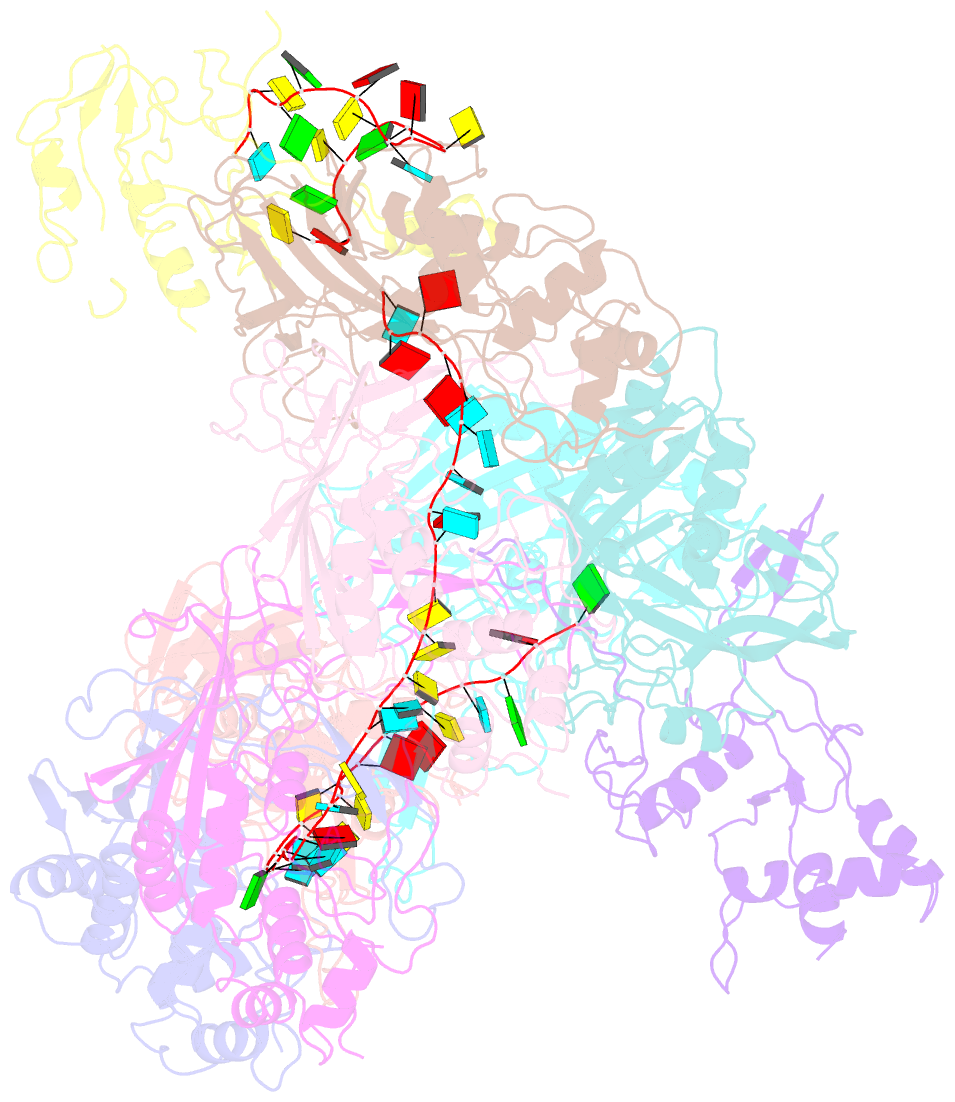
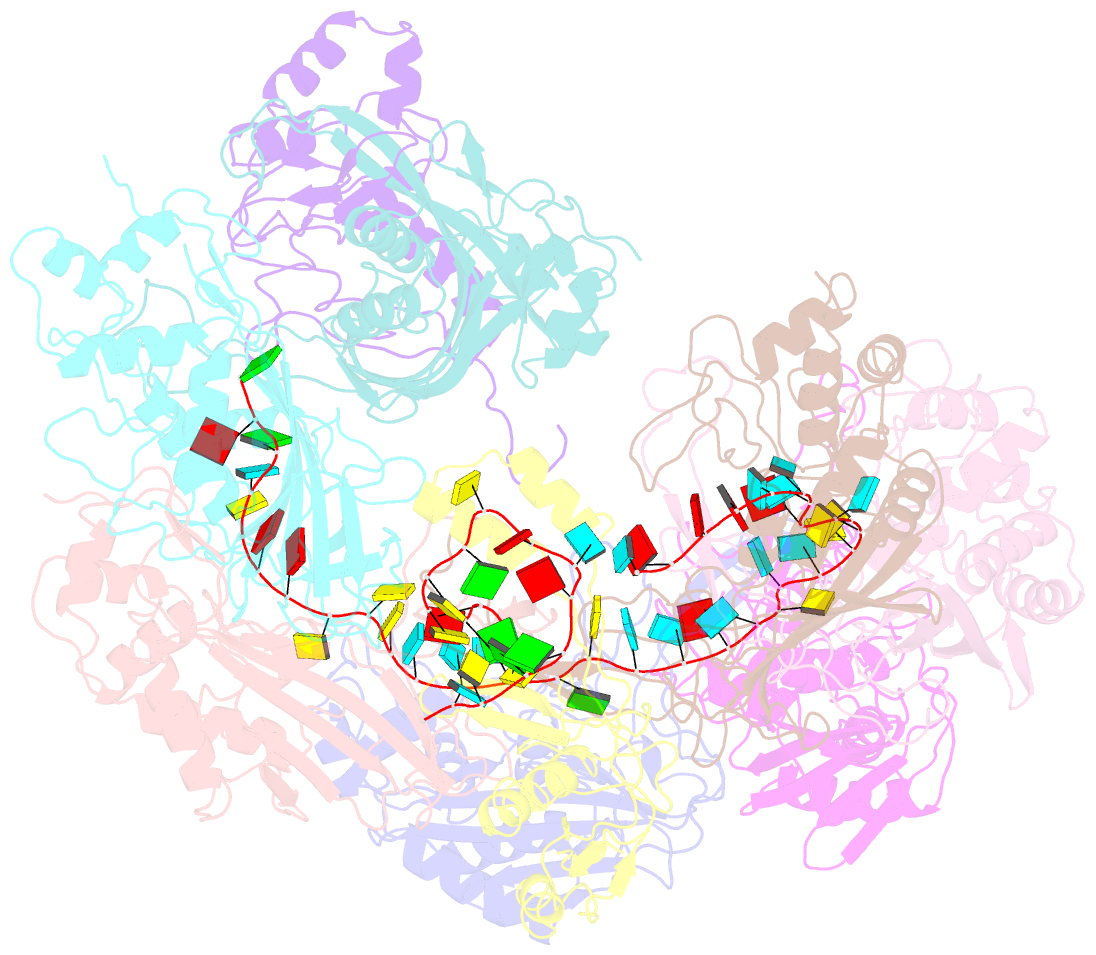
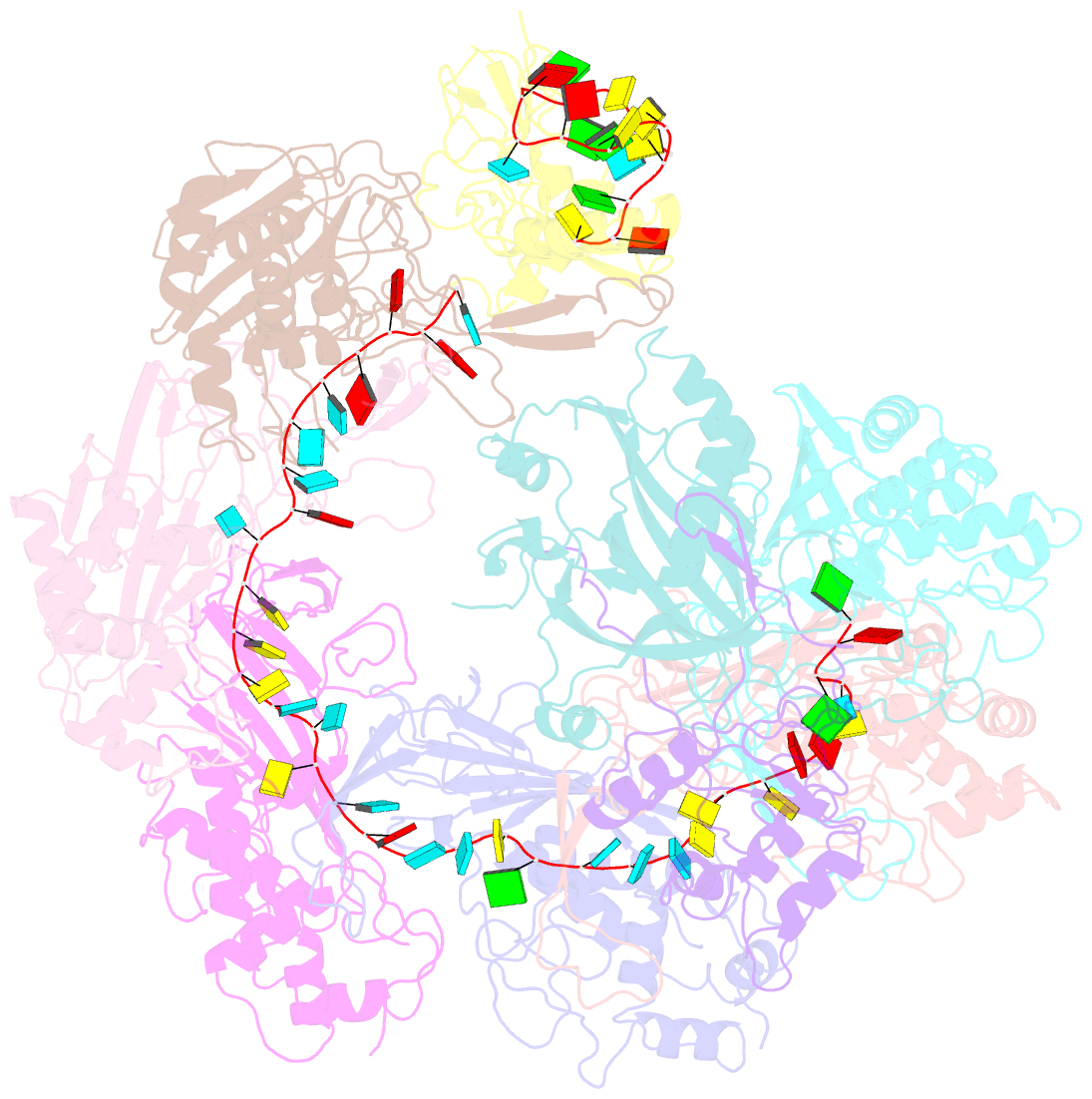
Summary information and primary citation
- PDB-id
- 7wwu; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- RNA binding protein-RNA
- Method
- cryo-EM (3.5 Å)
- Summary
- Icp1 csy complex
- Reference
- Zhang M, Peng R, Peng Q, Liu S, Li Z, Zhang Y, Song H, Yang J, Xing X, Wang P, Qi J, Gao GF (2023): "Mechanistic insights into DNA binding and cleavage by a compact type I-F CRISPR-Cas system in bacteriophage." Proc.Natl.Acad.Sci.USA, 120, e2215098120. doi: 10.1073/pnas.2215098120.
- Abstract
- CRISPR-Cas systems are widespread adaptive antiviral systems used in prokaryotes. Some phages, in turn, although have small genomes can economize the use of genetic space to encode compact or incomplete CRISPR-Cas systems to inhibit the host and establish infection. Phage ICP1, infecting Vibrio cholerae, encodes a compact type I-F CRISPR-Cas system to suppress the antiphage mobile genetic element in the host genome. However, the mechanism by which this compact system recognizes the target DNA and executes interference remains elusive. Here, we present the electron cryo-microscopy (cryo-EM) structures of both apo- and DNA-bound ICP1 surveillance complexes (Aka Csy complex). Unlike most other type I surveillance complexes, the ICP1 Csy complex lacks the Cas11 subunit or a structurally homologous domain, which is crucial for dsDNA binding and Cas3 activation in other type I CRISPR-Cas systems. Structural and functional analyses revealed that the compact ICP1 Csy complex alone is inefficient in binding to dsDNA targets, presumably stalled at a partial R-loop conformation. The presence of Cas2/3 facilitates dsDNA binding and allows effective dsDNA target cleavage. Additionally, we found that Pseudomonas aeruginosa Cas2/3 efficiently cleaved the dsDNA target presented by the ICP1 Csy complex, but not vice versa. These findings suggest a unique mechanism for target dsDNA binding and cleavage by the compact phage-derived CRISPR-Cas system.