Summary information and primary citation
- PDB-id
- 7x1n; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription-DNA
- Method
- X-ray (3.315 Å)
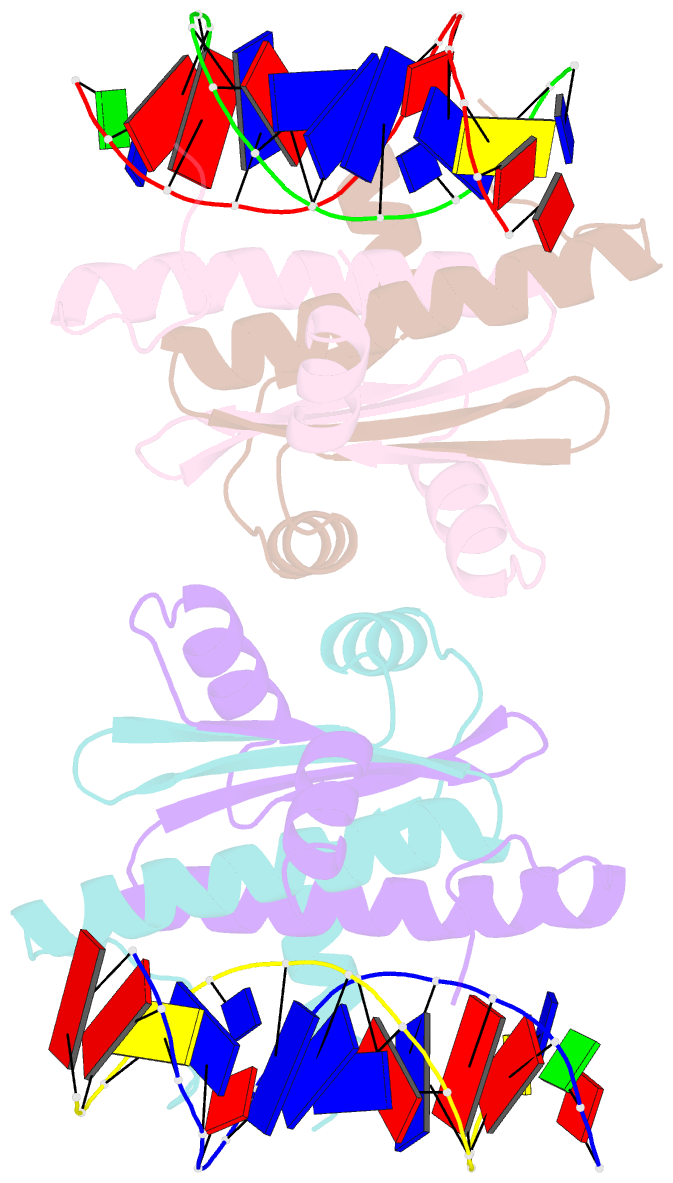
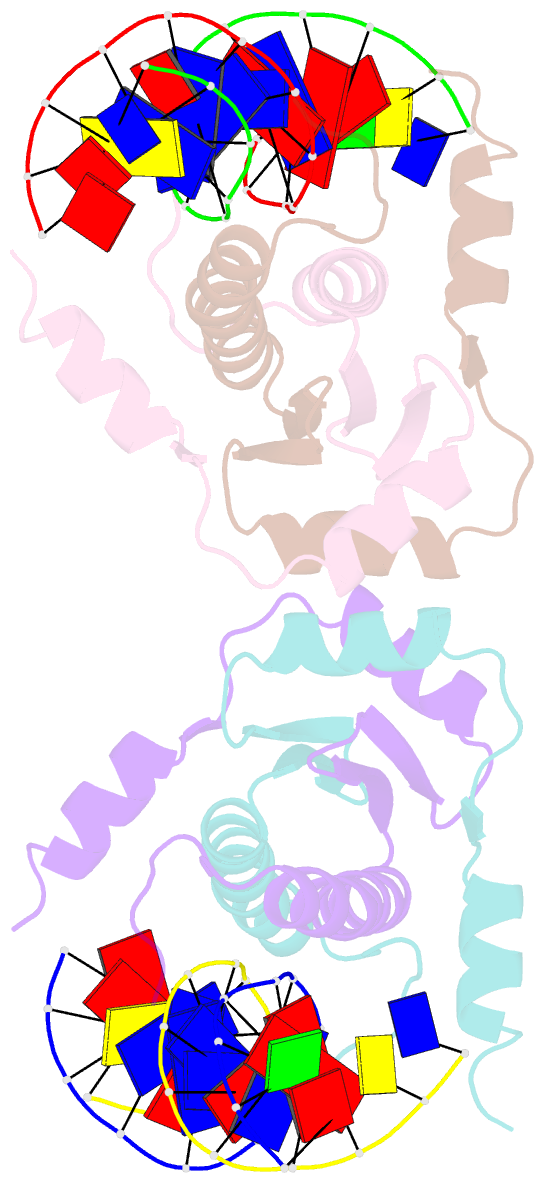
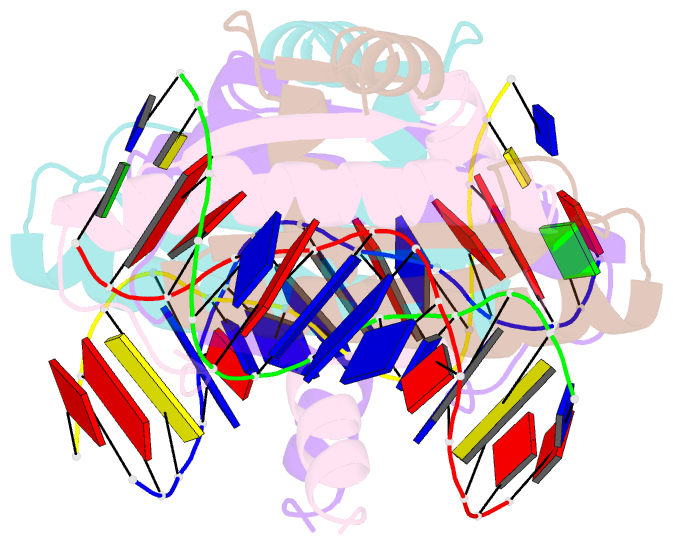
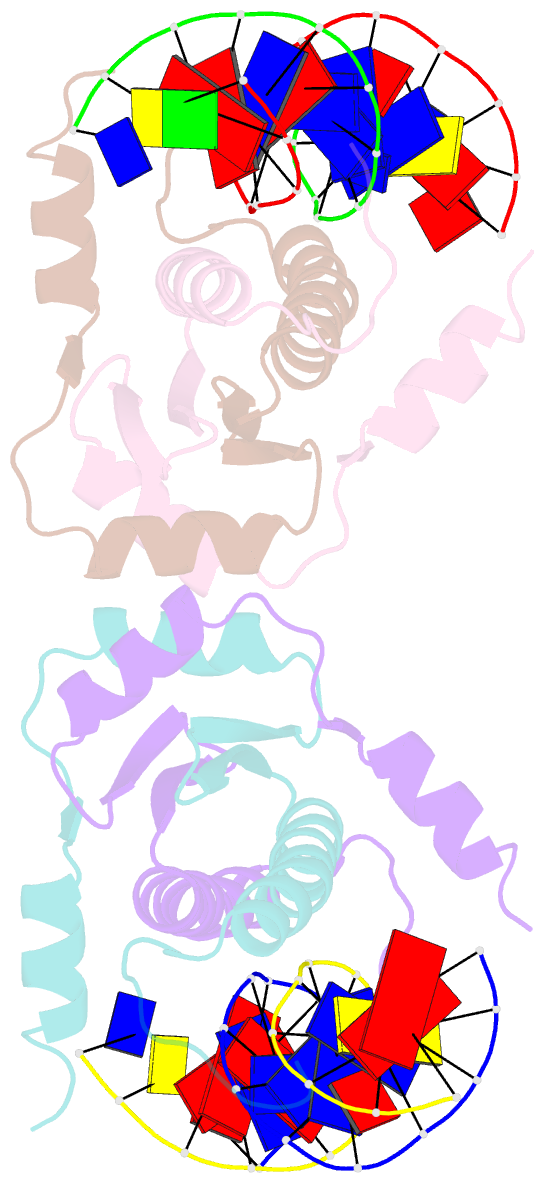
- Summary
- Crystal structure of mef2d-mre complex
- Reference
- Zhang M, Zhang H, Li Z, Bai L, Wang Q, Li J, Jiang M, Xue Q, Cheng N, Zhang W, Mao D, Chen Z, Huang J, Meng G, Chen Z, Chen SJ (2022): "Functional, structural, and molecular characterizations of the leukemogenic driver MEF2D-HNRNPUL1 fusion." Blood, 140, 1390-1407. doi: 10.1182/blood.2022016241.
- Abstract
- Recurrent MEF2D fusions with poor prognosis have been identified in B-cell precursor ALL (BCP-ALL). The molecular mechanisms underlying the pathogenic function of MEF2D fusions are poorly understood. Here, we showed that MEF2D-HNRNPUL1 (MH) knock-in mice developed a progressive disease from impaired B-cell development at pre-pro-B stage to pre-leukemia over 10 to 12 months. When cooperating with NRASG12D, MH drove an outbreak of BCP-ALL, with a more aggressive phenotype than the NRASG12D-induced leukemia. RNA-sequencing identified key networks involved in disease mechanisms. In ChIP-sequencing experiments, MH acquired increased chromatin-binding ability, mostly through MEF2D-Response-Element (MRE) motifs in target genes, compared to wild-type MEF2D. Using X-ray crystallography, the MEF2D-MRE complex was characterized in atomic resolution, while disrupting MH-DNA interaction alleviated the aberrant target gene expression and the B-cell differentiation arrest. The C-terminal moiety (HNRNPUL1 part) of MH was proven to contribute to the fusion protein's trans-regulatory activity, co-factors recruitment and homodimerization. Furthermore, targeting MH-driven transactivation of HDAC family using HDAC inhibitor Panobinostat improved the overall survival of MH/NRASG12D BCP-ALL mice in combination with chemotherapy. Altogether, these results not only highlight MH as an important driver in leukemogenesis, but also provoke targeted intervention against BCP-ALL with MEF2D fusions.