Summary information and primary citation
- PDB-id
- 7xug; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription
- Method
- cryo-EM (3.57 Å)
- Summary
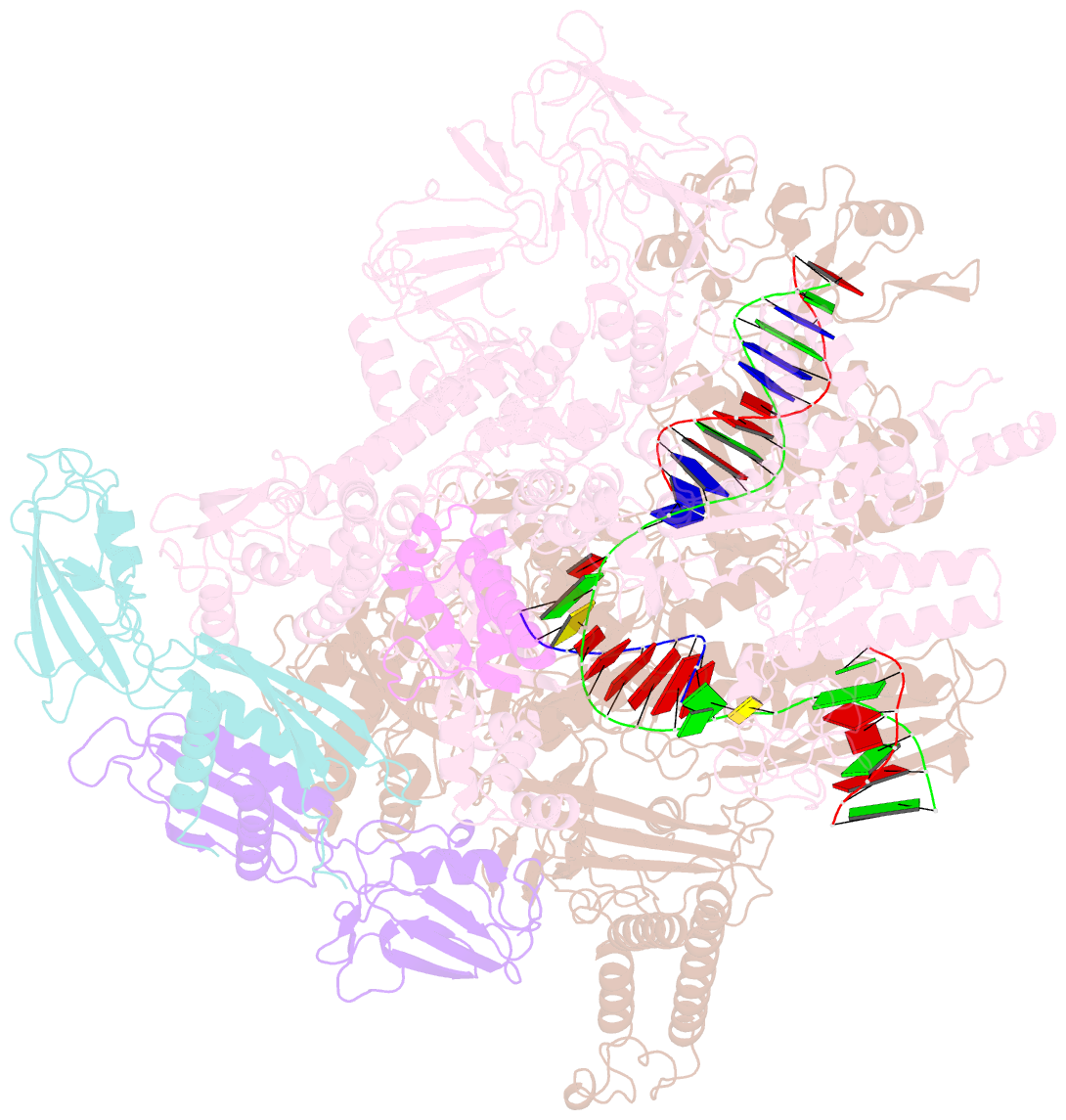
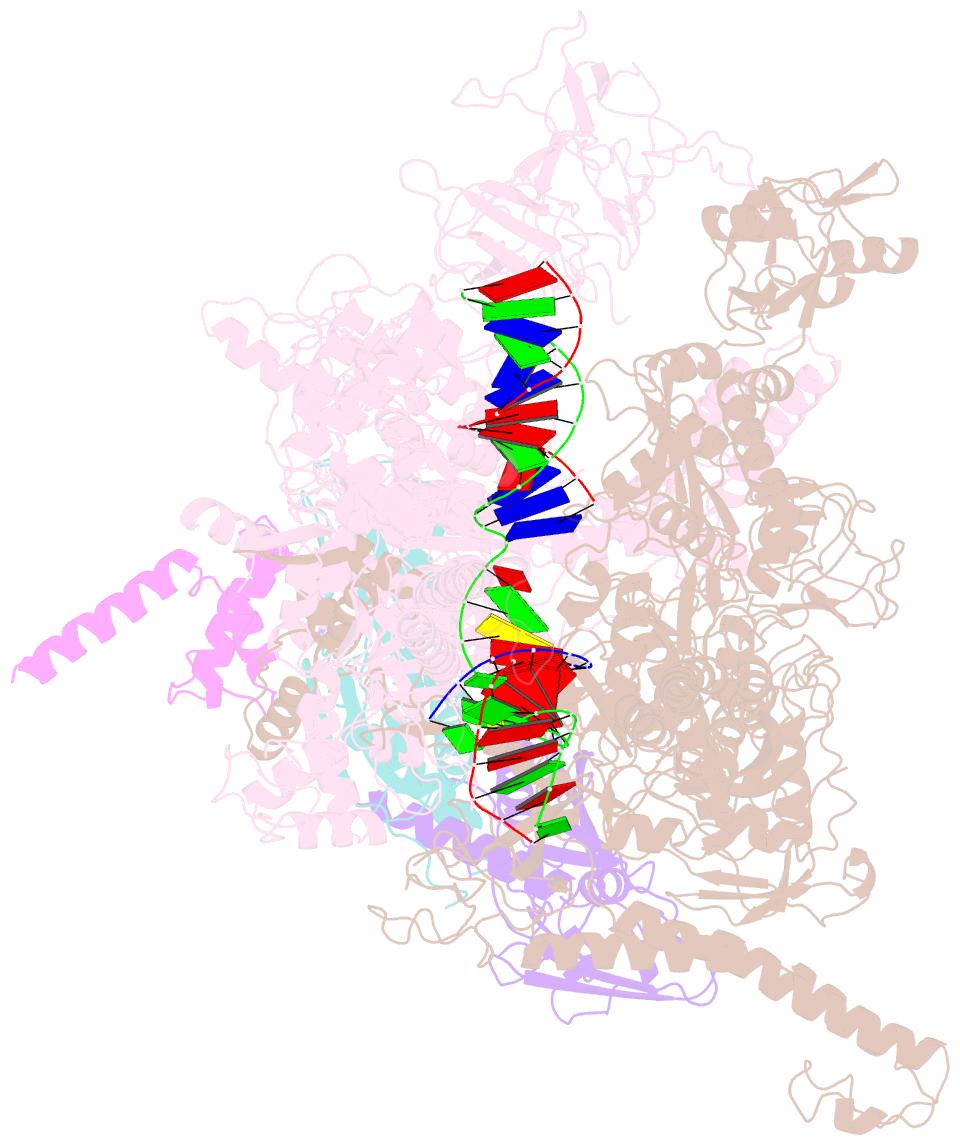
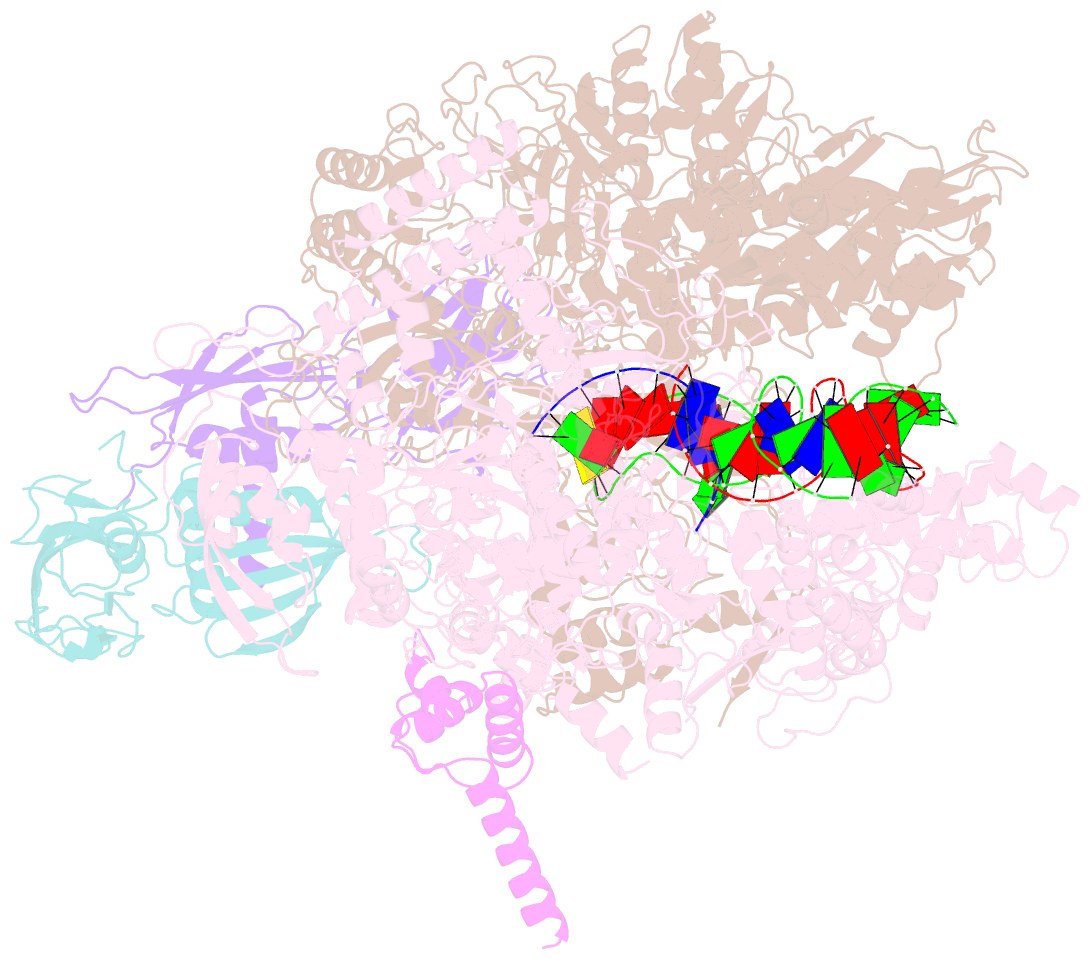
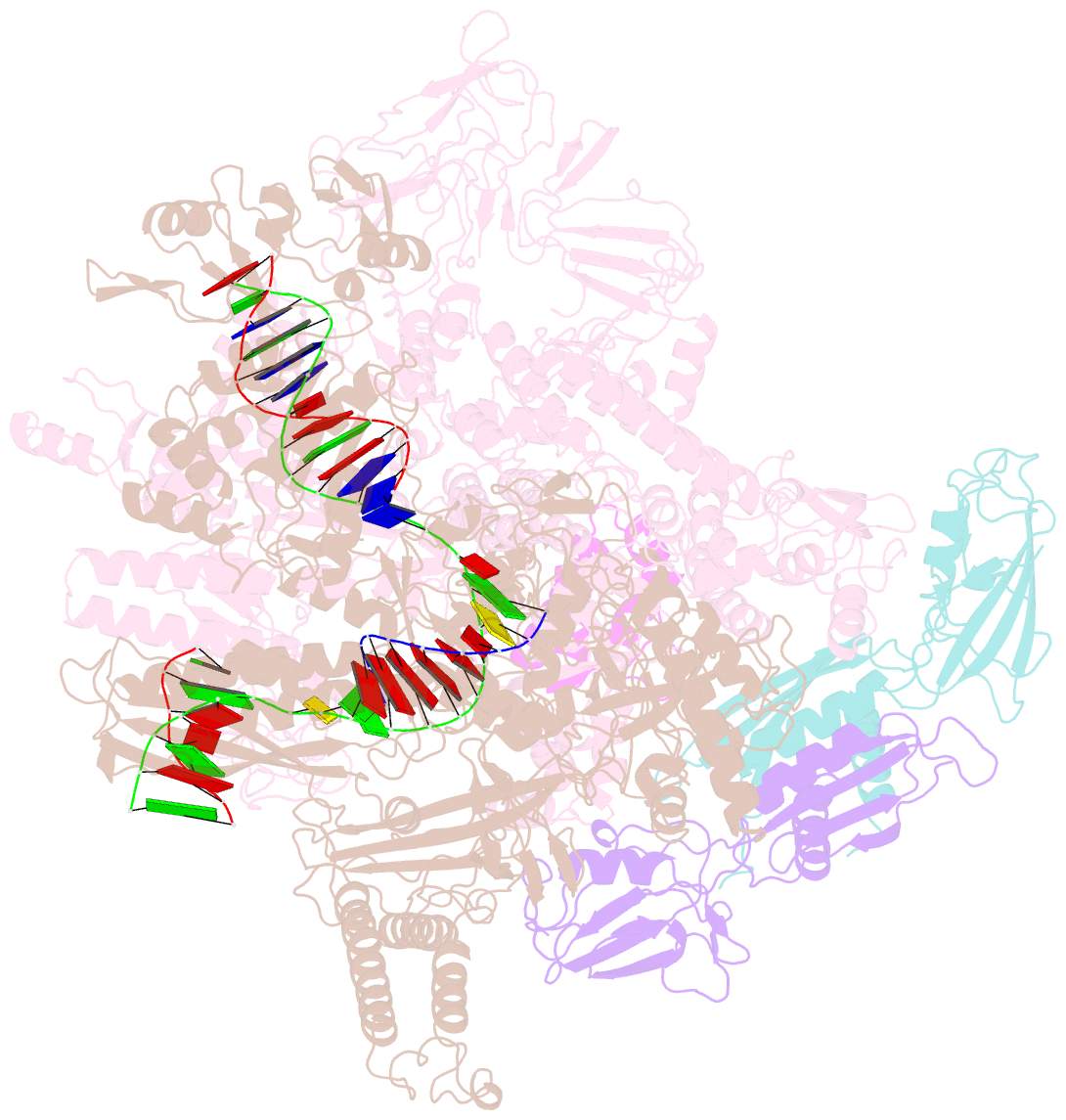
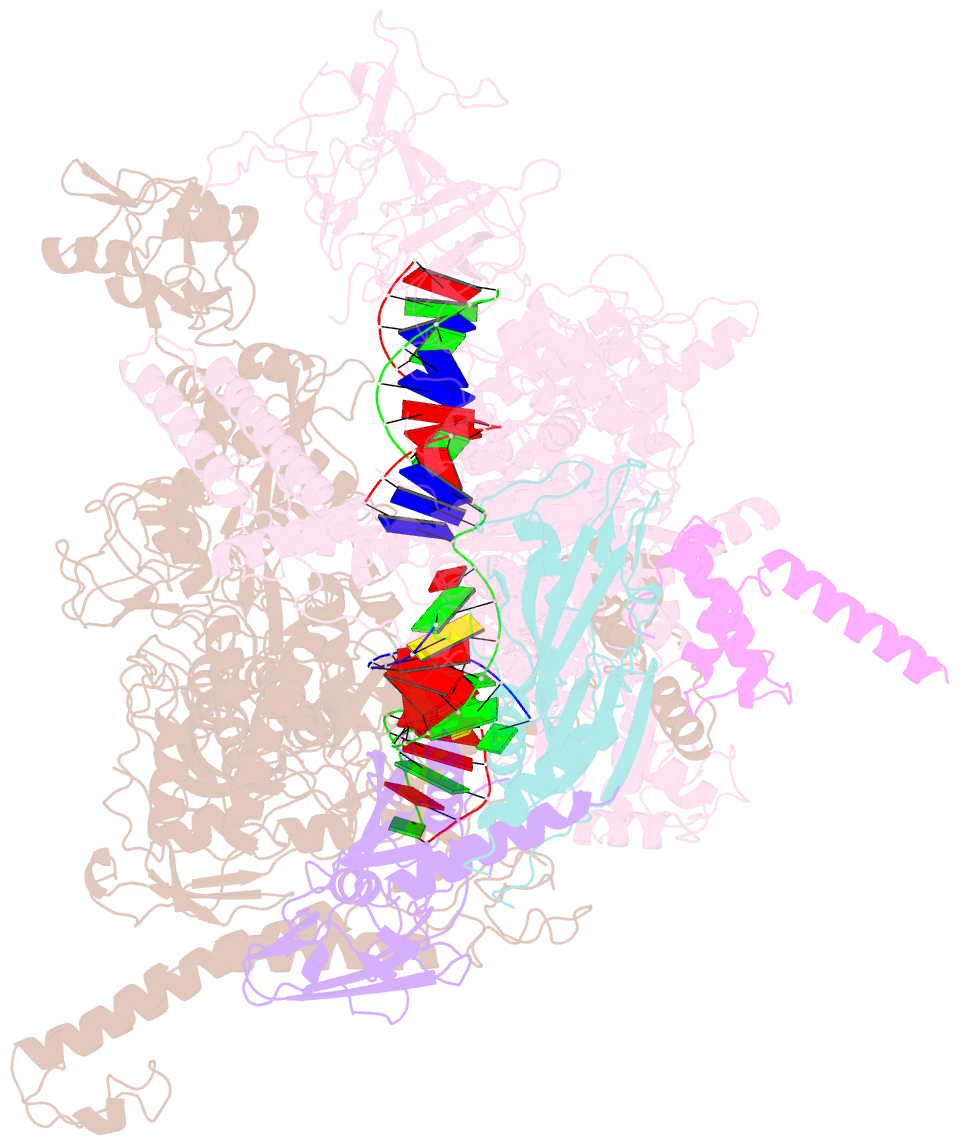
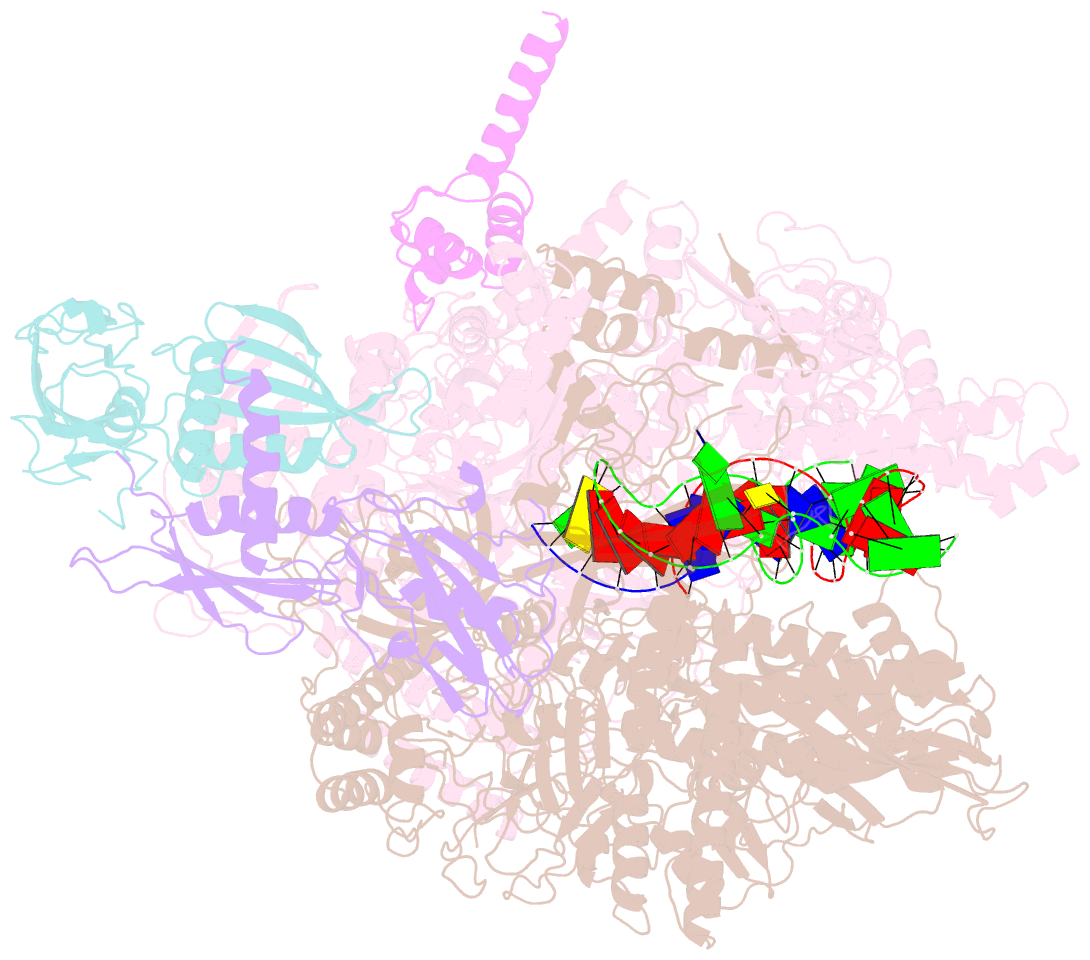
- cryo-EM structure of hk022 putrna-less e.coli RNA polymerase elongation complex
- Reference
- Hwang S, Olinares PDB, Lee J, Kim J, Chait BT, King RA, Kang JY (2022): "Structural basis of transcriptional regulation by a nascent RNA element, HK022 putRNA." Nat Commun, 13, 4668. doi: 10.1038/s41467-022-32315-y.
- Abstract
- Transcription, in which RNA polymerases (RNAPs) produce RNA from DNA, is the first step of gene expression. As such, it is highly regulated either by trans-elements like protein factors and/or by cis-elements like specific sequences on the DNA. Lambdoid phage HK022 contains a cis-element, put, which suppresses pausing and termination during transcription of the early phage genes. The putRNA transcript solely performs the anti-pausing/termination activities by interacting directly with the E.coli RNAP elongation complex (EC) by an unknown structural mechanism. In this study, we reconstituted putRNA-associated ECs and determined the structures using cryo-electron microscopy. The determined structures of putRNA-associated EC, putRNA-absent EC, and σ70-bound EC suggest that the putRNA interaction with the EC counteracts swiveling, a conformational change previously identified to promote pausing and σ70 might modulate putRNA folding via σ70-dependent pausing during elongation.