Summary information and primary citation
- PDB-id
- 7xuz; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription
- Method
- X-ray (3.591 Å)
- Summary
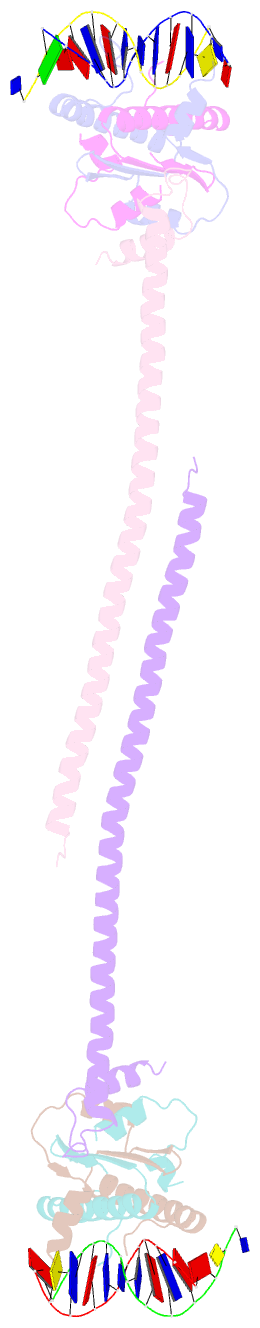
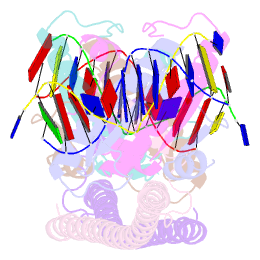
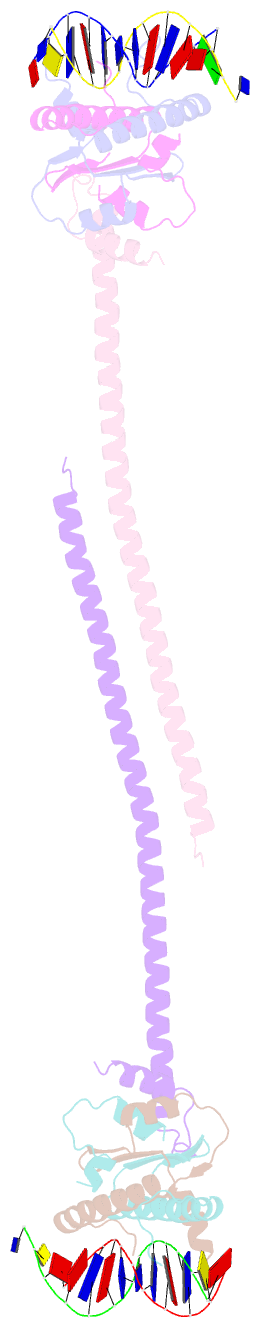
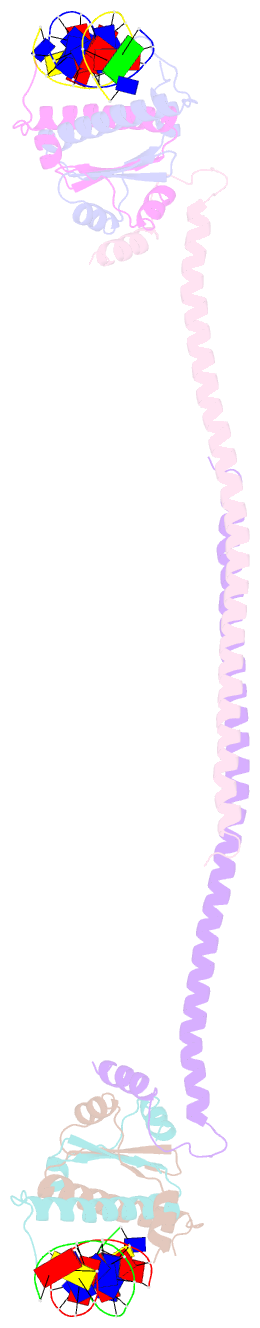
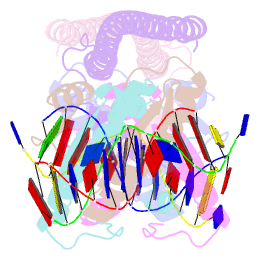
- Crystal structure of a hdac4-mef2a-DNA ternary complex
- Reference
- Dai S, Guo L, Dey R, Guo M, Zhang X, Bates D, Cayford J, Jiang L, Wei H, Chen Z, Zhang Y, Chen L, Chen Y (2024): "Structural insights into the HDAC4-MEF2A-DNA complex and its implication in long-range transcriptional regulation." Nucleic Acids Res., 52, 2711-2723. doi: 10.1093/nar/gkae036.
- Abstract
- Class IIa Histone deacetylases (HDACs), including HDAC4, 5, 7 and 9, play key roles in multiple important developmental and differentiation processes. Recent studies have shown that class IIa HDACs exert their transcriptional repressive function by interacting with tissue-specific transcription factors, such as members of the myocyte enhancer factor 2 (MEF2) family of transcription factors. However, the molecular mechanism is not well understood. In this study, we determined the crystal structure of an HDAC4-MEF2A-DNA complex. This complex adopts a dumbbell-shaped overall architecture, with a 2:4:2 stoichiometry of HDAC4, MEF2A and DNA molecules. In the complex, two HDAC4 molecules form a dimer through the interaction of their glutamine-rich domain (GRD) to form the stem of the 'dumbbell'; while two MEF2A dimers and their cognate DNA molecules are bridged by the HDAC4 dimer. Our structural observations were then validated using biochemical and mutagenesis assays. Further cell-based luciferase reporter gene assays revealed that the dimerization of HDAC4 is crucial in its ability to repress the transcriptional activities of MEF2 proteins. Taken together, our findings not only provide the structural basis for the assembly of the HDAC4-MEF2A-DNA complex but also shed light on the molecular mechanism of HDAC4-mediated long-range gene regulation.