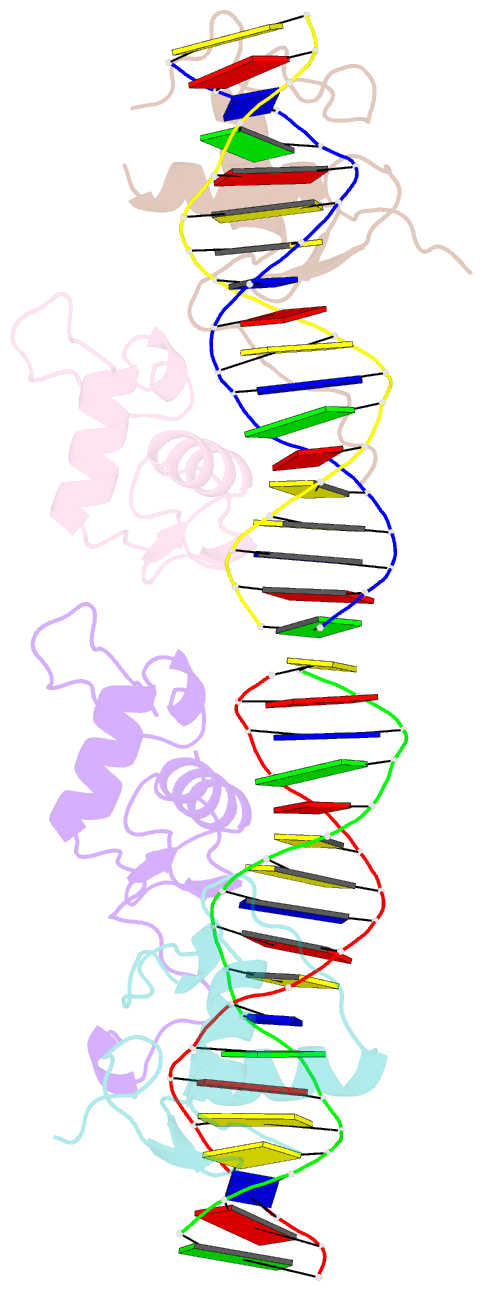
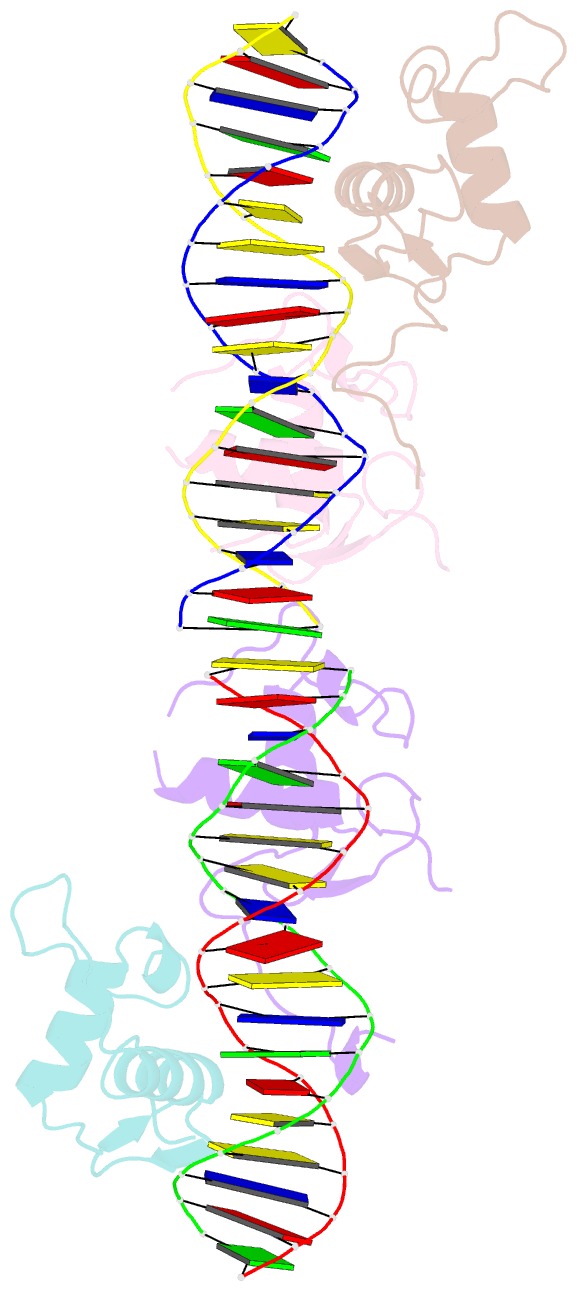
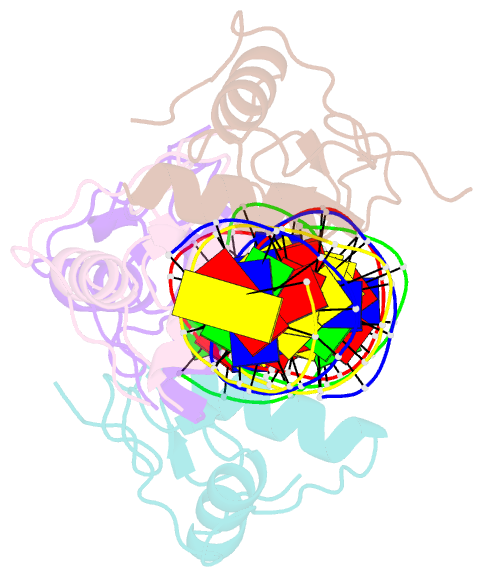
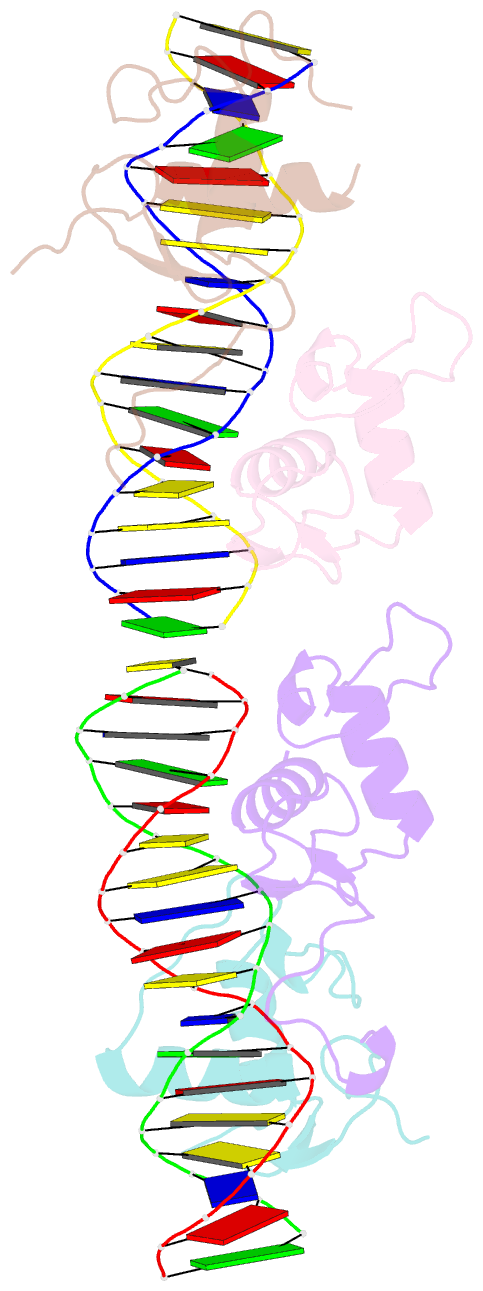
Summary information and primary citation
- PDB-id
- 7xvn; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- DNA binding protein-DNA
- Method
- X-ray (2.302 Å)
- Summary
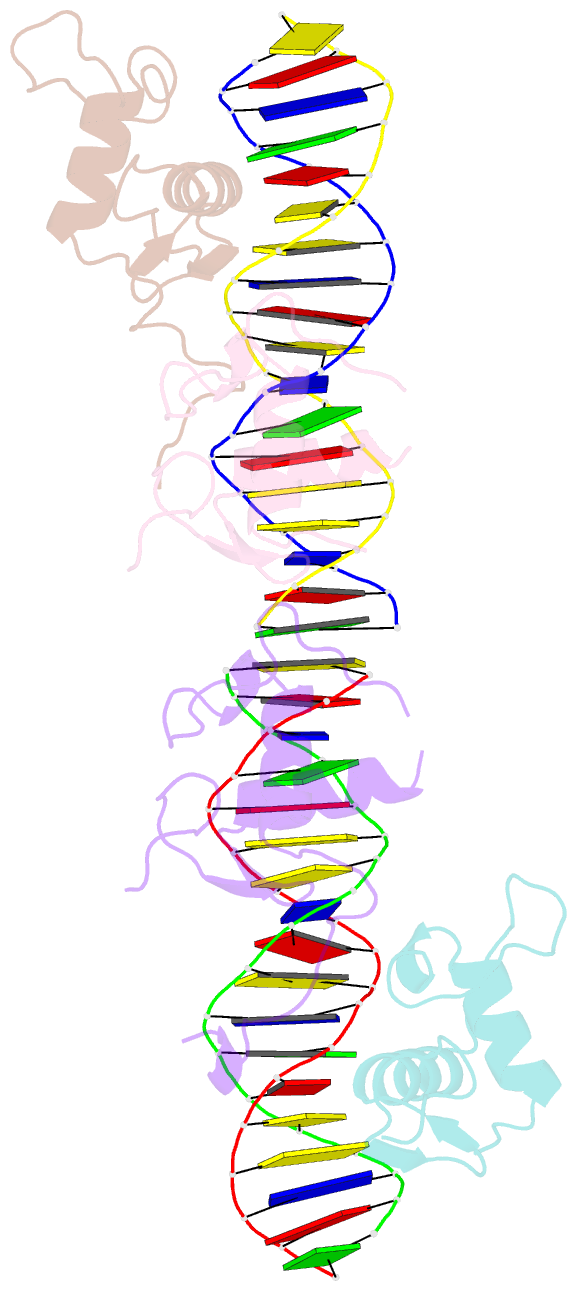
- Structural basis for DNA recognition feature of retinoid-related orphan receptors
- Reference
- Jiang L, Liu X, Liang X, Dai S, Wei H, Guo M, Chen Z, Xiao D, Chen Y (2024): "Structural characterization of the DNA binding mechanism of retinoic acid-related orphan receptor gamma." Structure, 32, 467-475.e3. doi: 10.1016/j.str.2024.01.004.
- Abstract
- Retinoic acid-related orphan receptor gamma (RORγ) plays critical roles in regulating various biological processes and has been linked to immunodeficiency disorders and cancers. DNA recognition is essential for RORγ to exert its functions. However, the underlying mechanism of the DNA binding by RORγ remains unclear. In this study, we present the crystal structure of the complex of RORγ1 DNA-binding domain (RORγ1-DBD)/direct repeat DNA element DR2 at 2.3 Å resolution. We demonstrate that RORγ1-DBD binds the DR2 motif as a homodimer, with the C-terminal extension (CTE) region of RORγ1-DBD contributing to the DNA recognition and the formation of dimeric interface. Further studies reveal that REV-ERB-DBD and RXR-DBD, also bind the DR2 site as a homodimer, while NR4A2-DBD binds DR2 as a monomer. Our research uncovers a binding mechanism of RORγ1 to the DR2 site and provides insights into the biological functions of RORγ1 and the broader RORs subfamily.