Summary information and primary citation
- PDB-id
- 7xwz; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- viral protein-DNA
- Method
- X-ray (2.25 Å)
- Summary
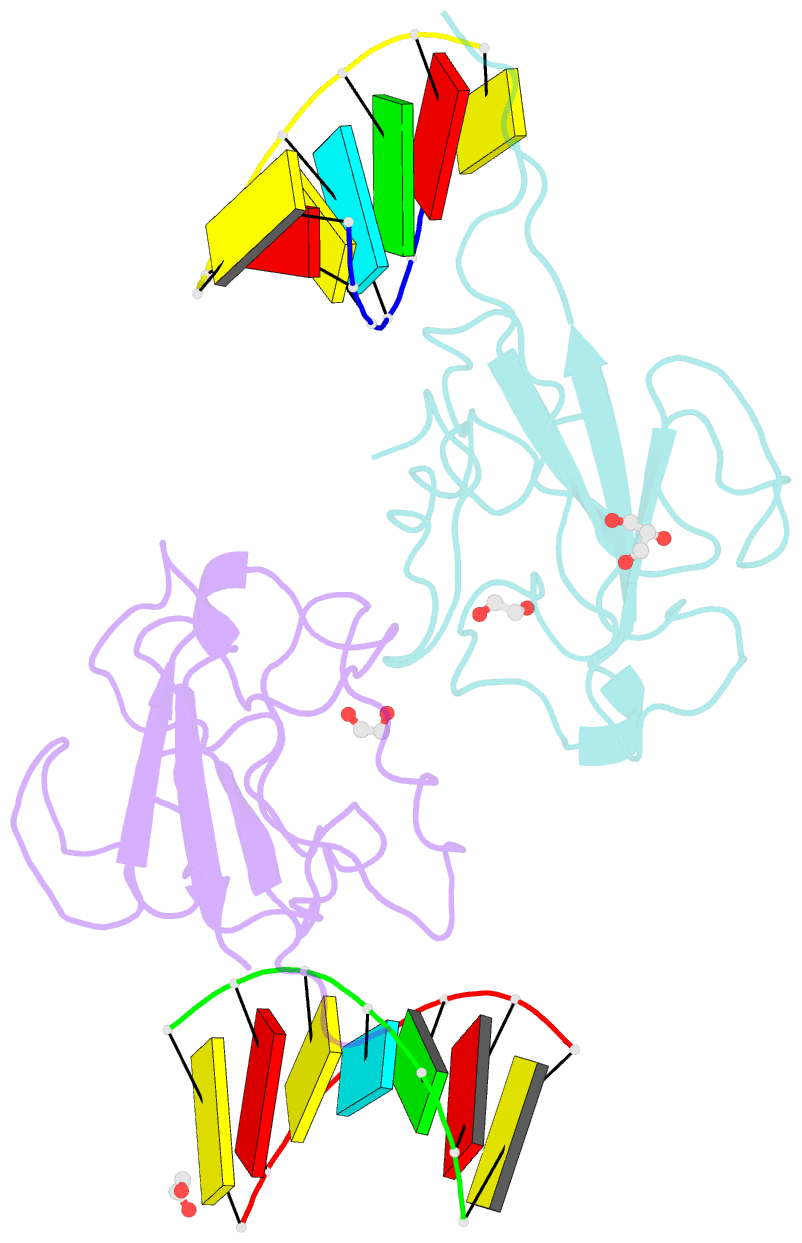
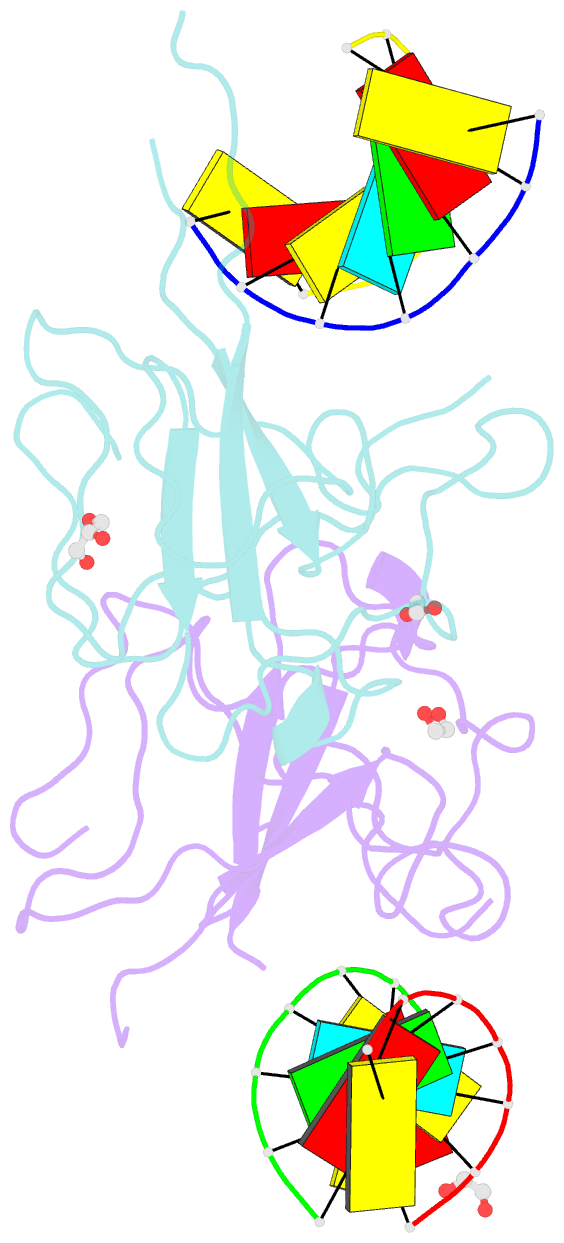
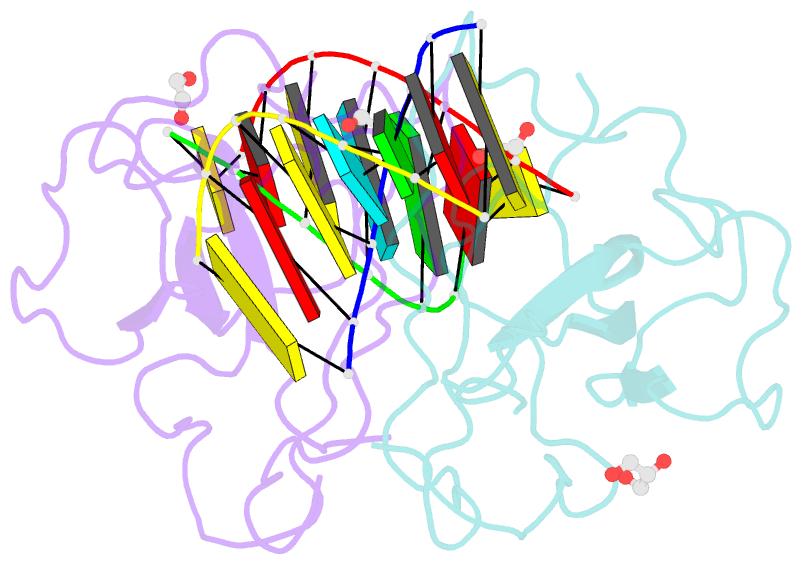
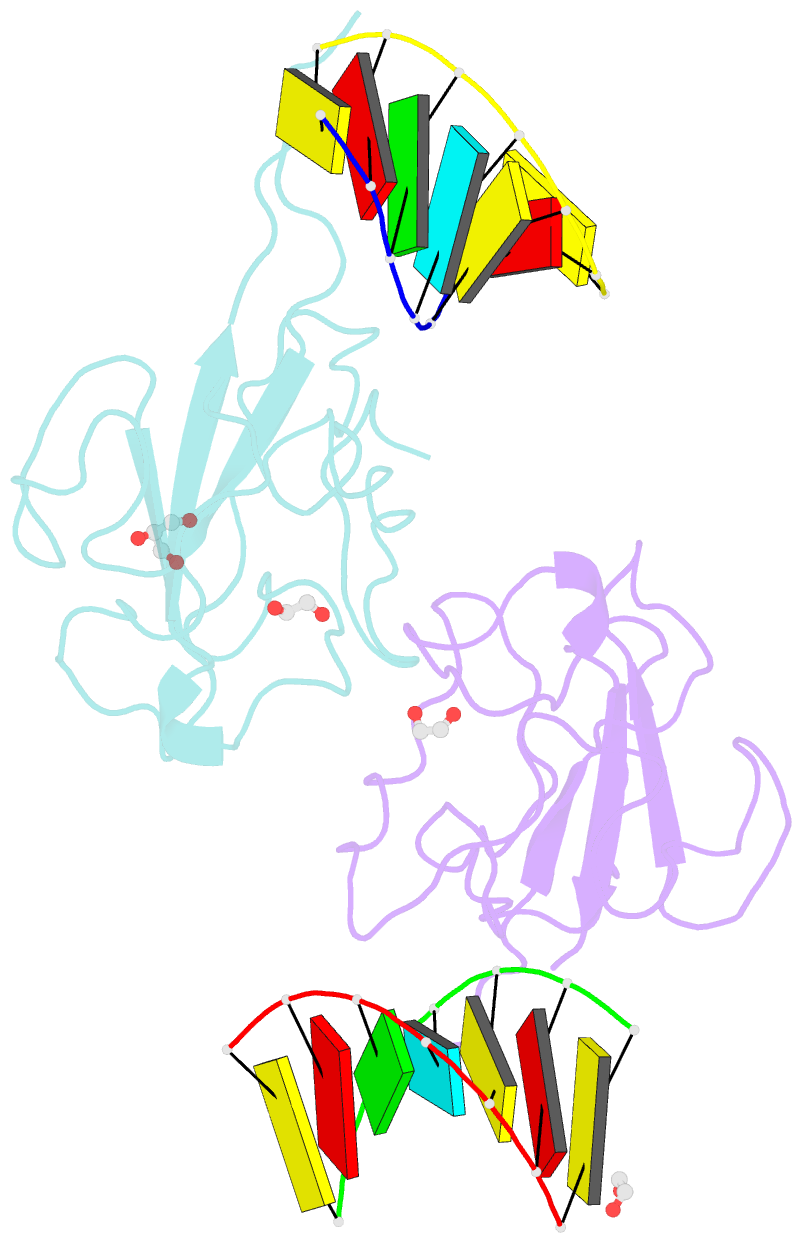
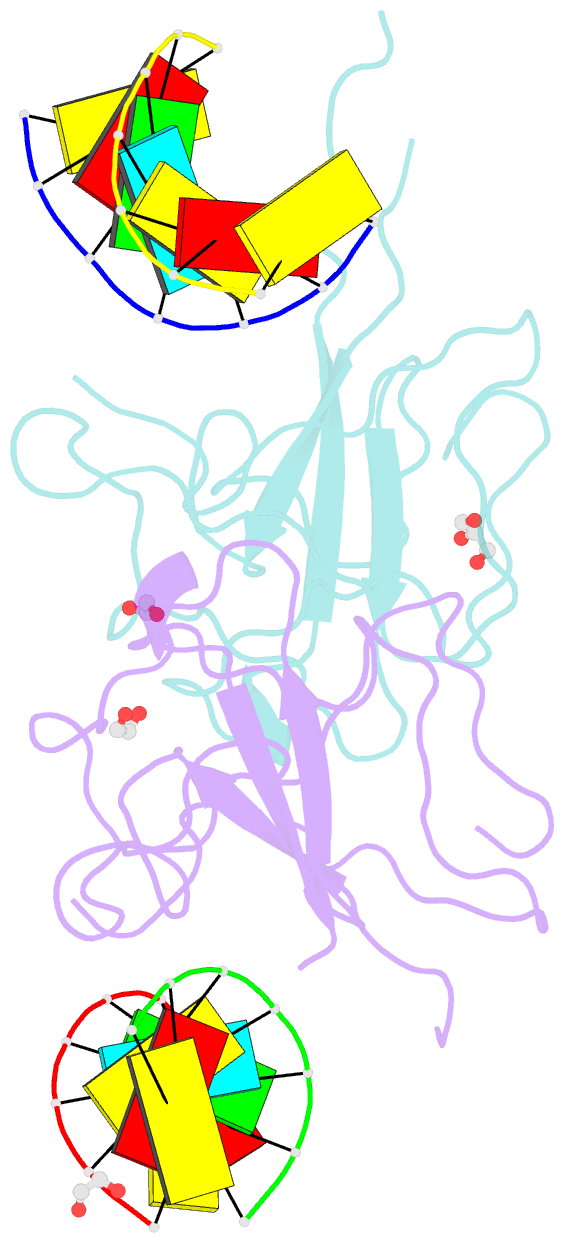
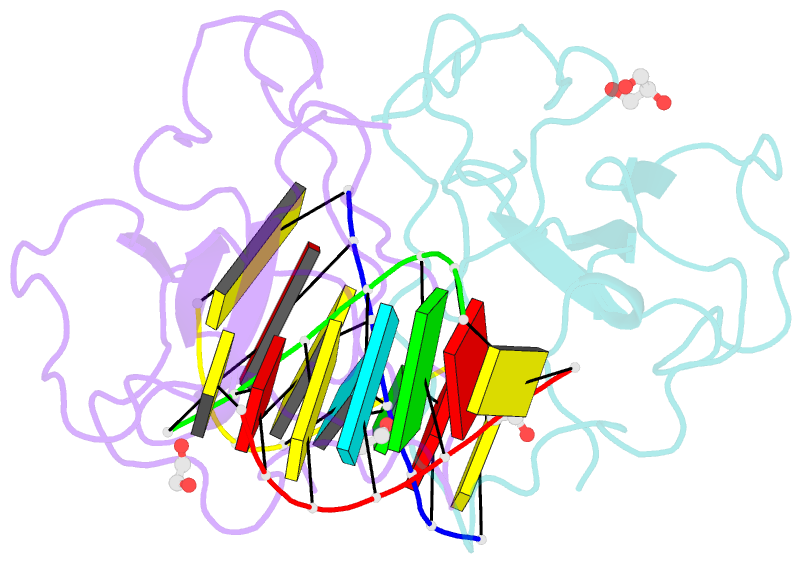
- Crystal structure of sars-cov-2 n-ntd and dsrna complex
- Reference
- Luan X, Li X, Li Y, Su G, Yin W, Jiang Y, Xu N, Wang F, Cheng W, Jin Y, Zhang L, Xu HE, Xue Y, Zhang S (2022): "Antiviral drug design based on structural insights into the N-terminal domain and C-terminal domain of the SARS-CoV-2 nucleocapsid protein." Sci Bull (Beijing), 67, 2327-2335. doi: 10.1016/j.scib.2022.10.021.
- Abstract
- Nucleocapsid (N) protein plays crucial roles in the life cycle of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), including the formation of ribonucleoprotein (RNP) complex with the viral RNA. Here we reported the crystal structures of the N-terminal domain (NTD) and C-terminal domain (CTD) of the N protein and an NTD-RNA complex. Our structures reveal a unique tetramer organization of NTD and identify a distinct RNA binding mode in the NTD-RNA complex, which could contribute to the formation of the RNP complex. We also screened small molecule inhibitors of N-NTD and N-CTD and discovered that ceftriaxone sodium, an antibiotic, can block the binding of RNA to NTD and inhibit the formation of the RNP complex. These results together could facilitate the further research of antiviral drug design targeting N protein.