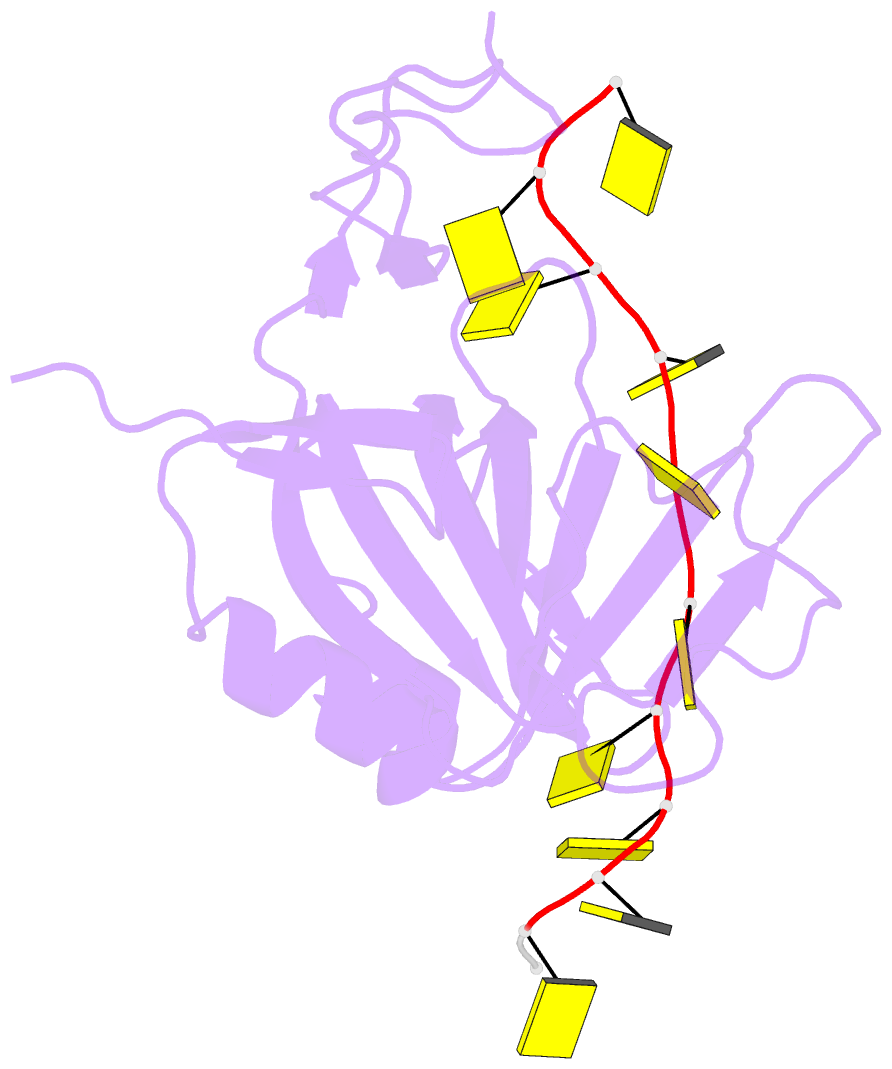
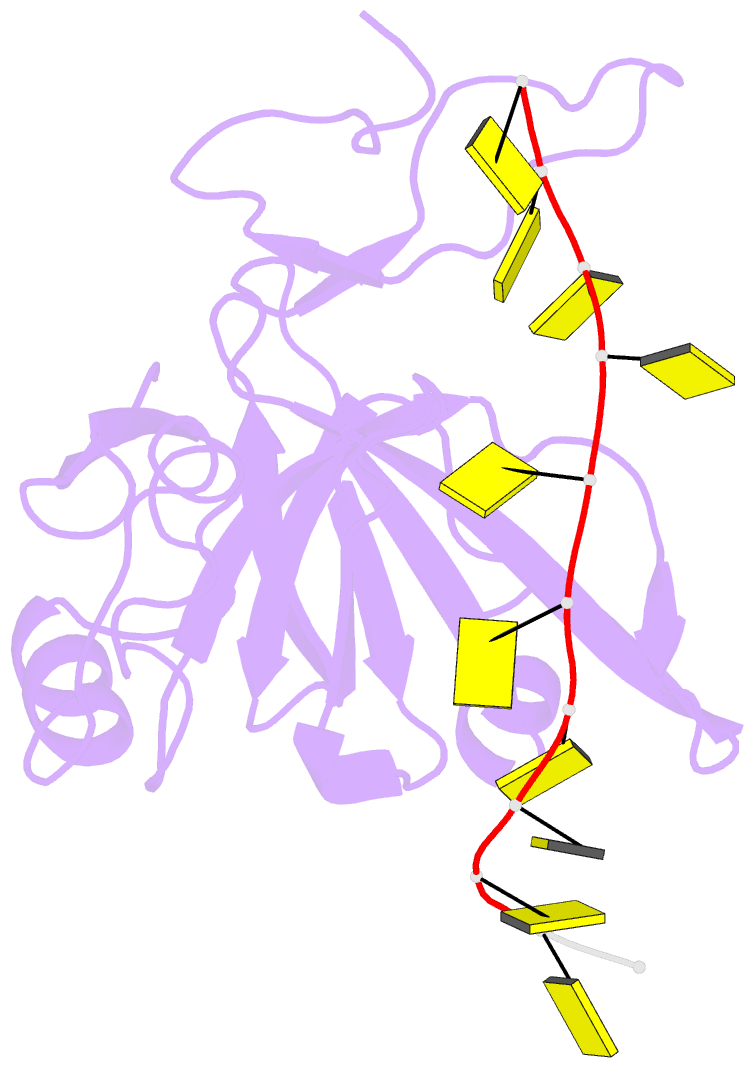
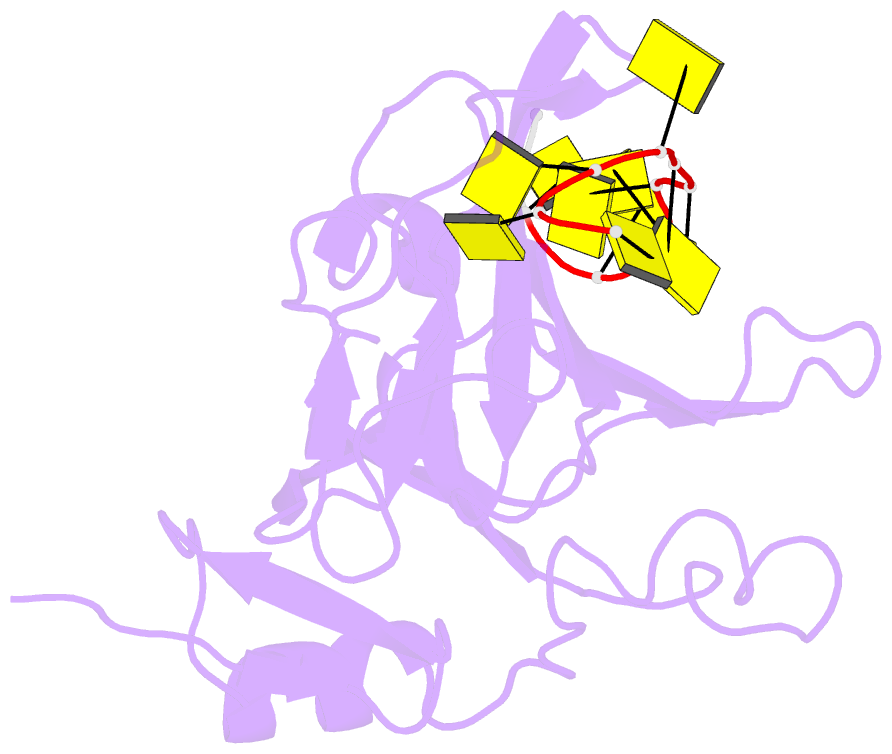
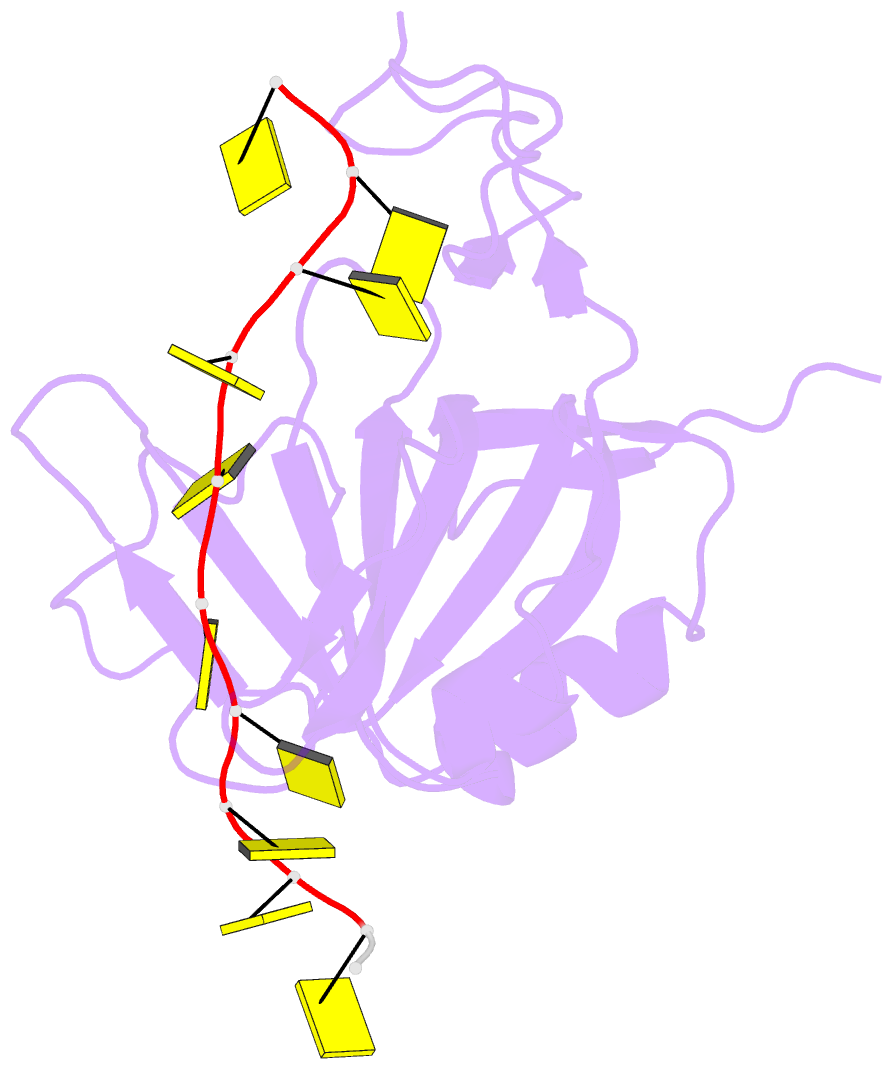
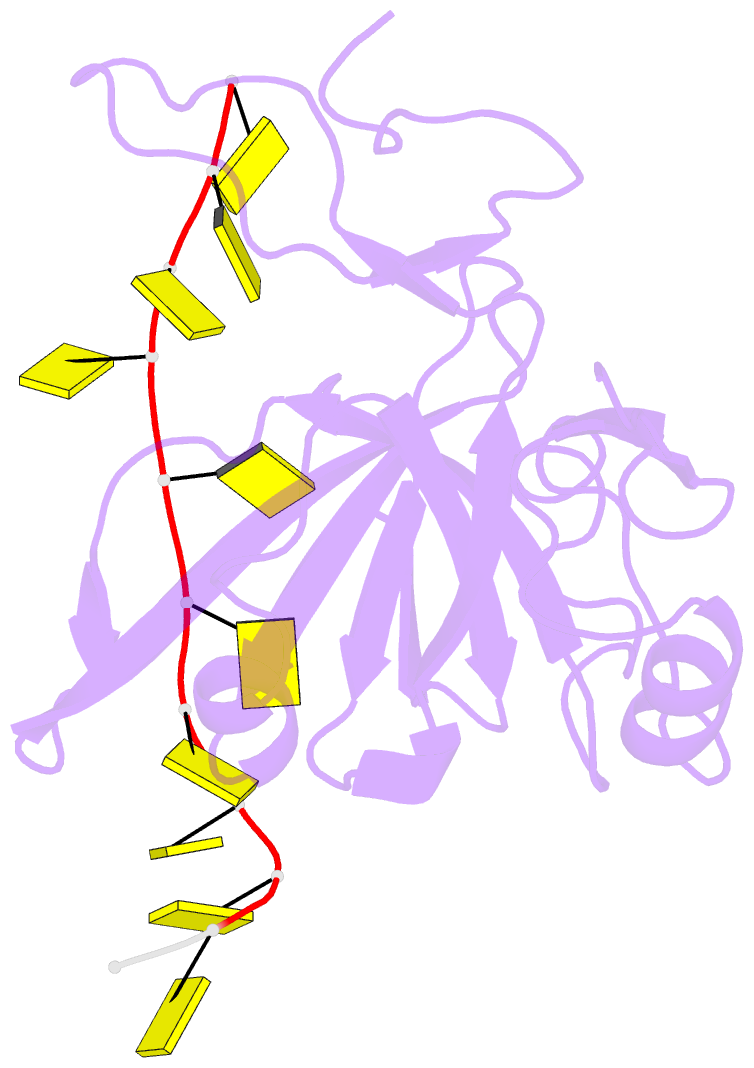
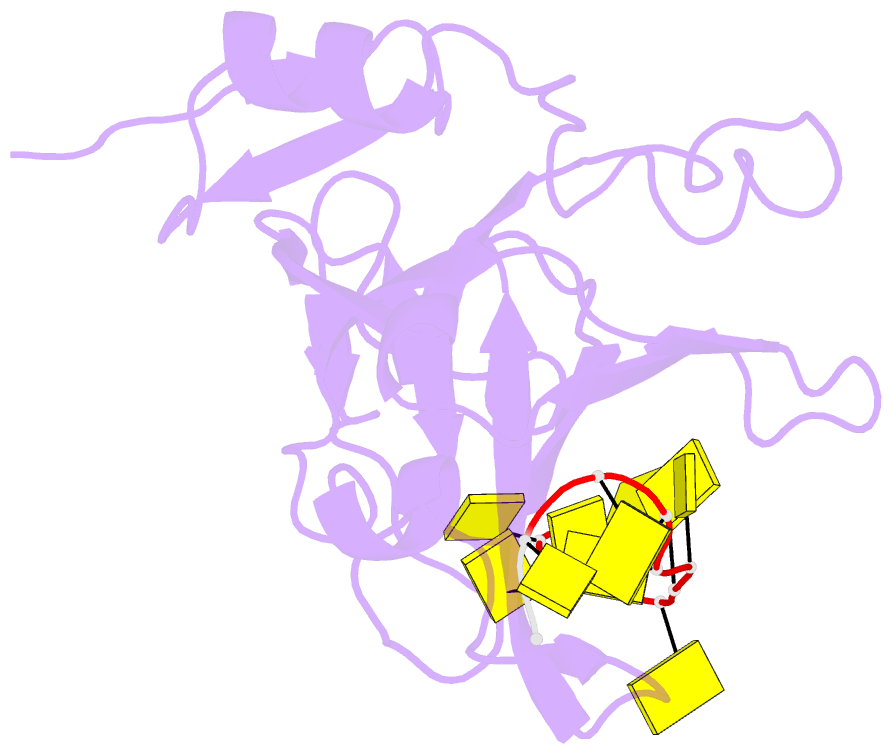
Summary information and primary citation
- PDB-id
- 7y01; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- plant protein-DNA
- Method
- X-ray (2.8 Å)
- Summary
- Crystal structure of zmmcm10 in complex with 16nt ssDNA at 2.8. angstrom resolution
- Reference
- Zhao X, Wang J, Jin D, Cheng J, Chen H, Li Z, Wang Y, Lou H, Zhu JK, Du X, Gong Z (2023): "AtMCM10 promotes DNA replication-coupled nucleosome assembly in Arabidopsis." J Integr Plant Biol, 65, 203-222. doi: 10.1111/jipb.13438.
- Abstract
- Minichromosome Maintenance protein 10 (MCM10) is essential for DNA replication initiation and DNA elongation in yeasts and animals. Although the functions of MCM10 in DNA replication and repair have been well documented, the detailed mechanisms for MCM10 in these processes are not well known. Here, we identified AtMCM10 gene through a forward genetic screening for releasing a silenced marker gene. Although plant MCM10 possesses a similar crystal structure as animal MCM10, AtMCM10 is not essential for plant growth or development in Arabidopsis. AtMCM10 can directly bind to histone H3-H4 and promotes nucleosome assembly in vitro. The nucleosome density is decreased in Atmcm10, and most of the nucleosome density decreased regions in Atmcm10 are also regulated by newly synthesized histone chaperone Chromatin Assembly Factor-1 (CAF-1). Loss of both AtMCM10 and CAF-1 is embryo lethal, indicating that AtMCM10 and CAF-1 are indispensable for replication-coupled nucleosome assembly. AtMCM10 interacts with both new and parental histones. Atmcm10 mutants have lower H3.1 abundance and reduced H3K27me1/3 levels with releasing some silenced transposons. We propose that AtMCM10 deposits new and parental histones during nucleosome assembly, maintaining proper epigenetic modifications and genome stability during DNA replication.