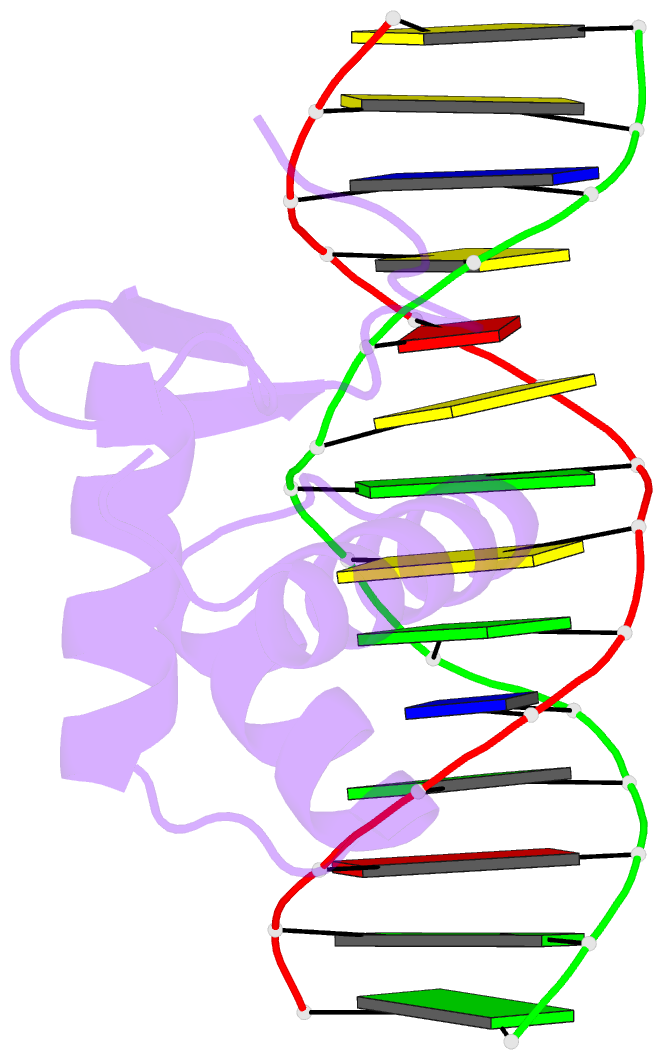
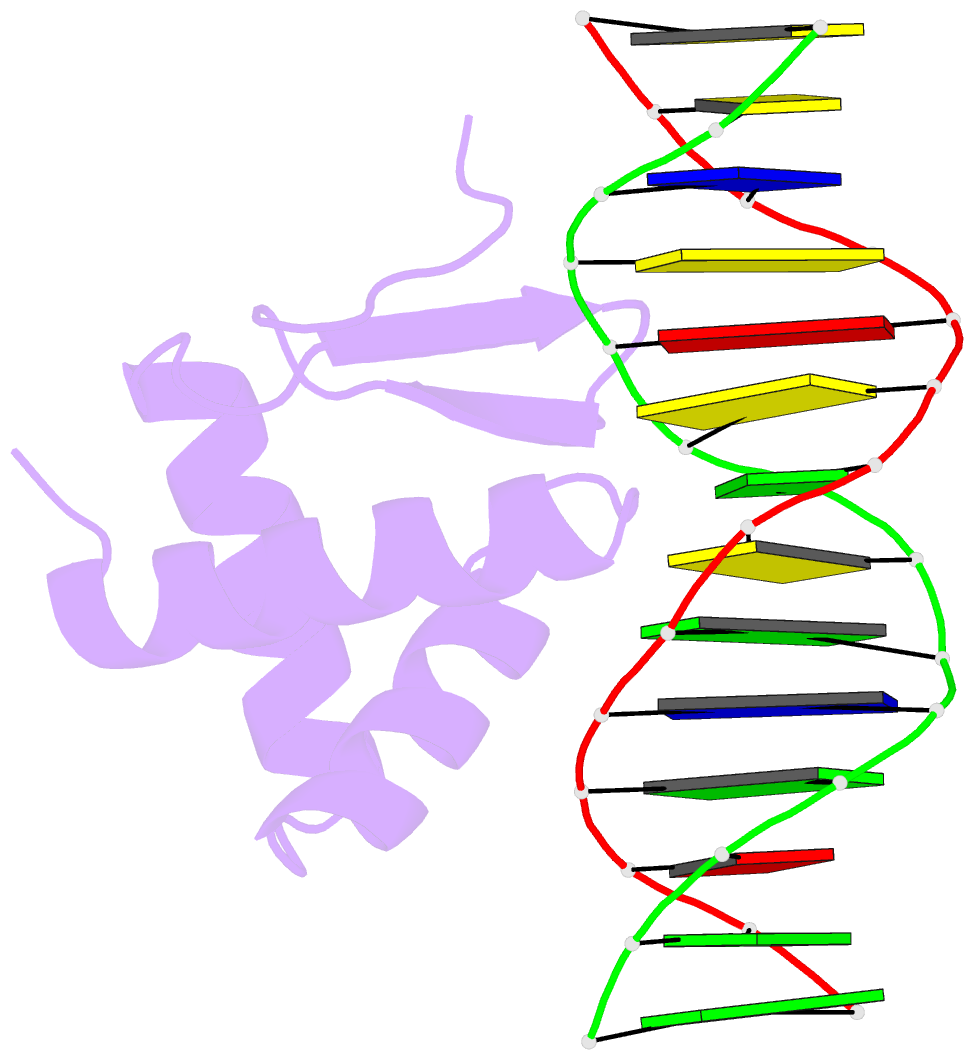
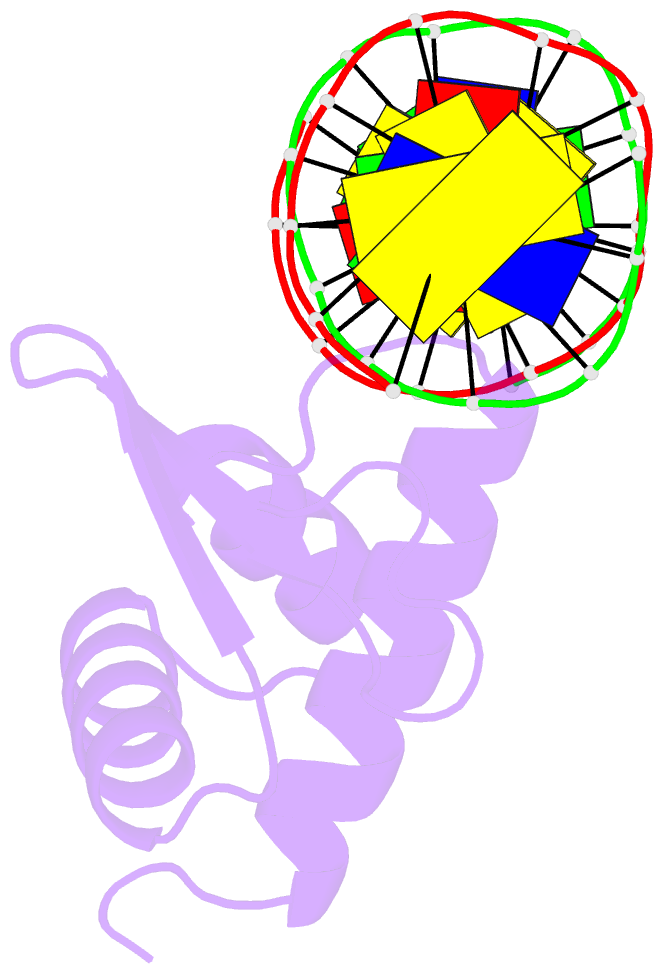
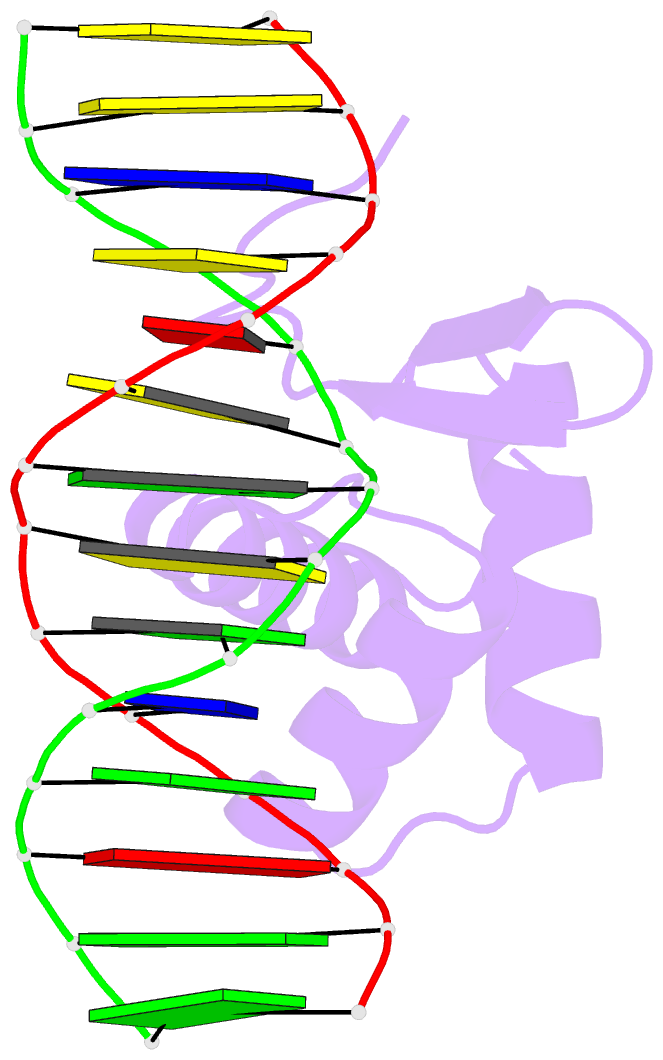
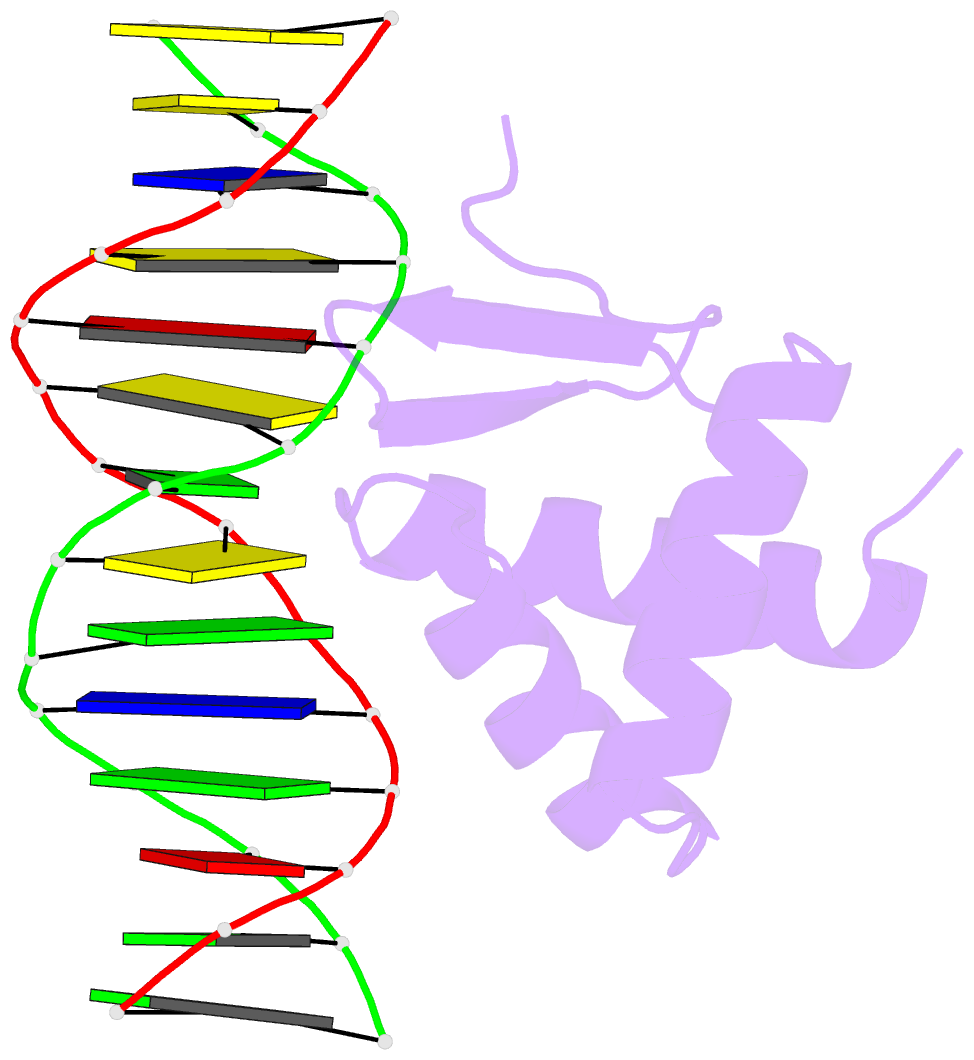
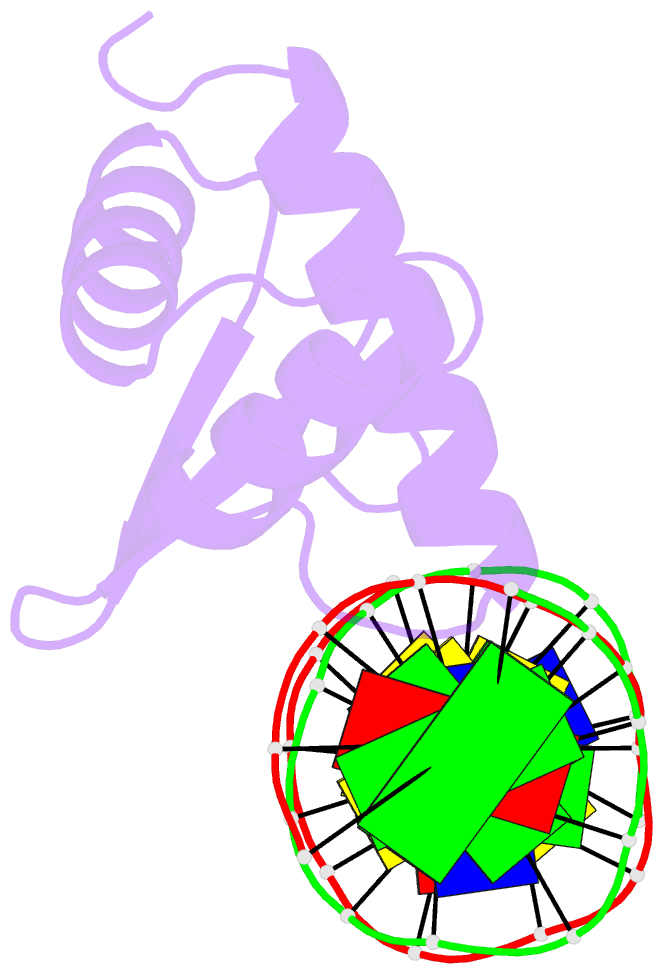
Summary information and primary citation
- PDB-id
- 7y43; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- DNA binding protein-DNA
- Method
- X-ray (1.5 Å)
- Summary
- Crystal structure of the kat6a wh domain and its bound double stranded DNA
- Reference
- Weber LM, Jia Y, Stielow B, Gisselbrecht SS, Cao Y, Ren Y, Rohner I, King J, Rothman E, Fischer S, Simon C, Forne I, Nist A, Stiewe T, Bulyk ML, Wang Z, Liefke R (2023): "The histone acetyltransferase KAT6A is recruited to unmethylated CpG islands via a DNA binding winged helix domain." Nucleic Acids Res., 51, 574-594. doi: 10.1093/nar/gkac1188.
- Abstract
- The lysine acetyltransferase KAT6A (MOZ, MYST3) belongs to the MYST family of chromatin regulators, facilitating histone acetylation. Dysregulation of KAT6A has been implicated in developmental syndromes and the onset of acute myeloid leukemia (AML). Previous work suggests that KAT6A is recruited to its genomic targets by a combinatorial function of histone binding PHD fingers, transcription factors and chromatin binding interaction partners. Here, we demonstrate that a winged helix (WH) domain at the very N-terminus of KAT6A specifically interacts with unmethylated CpG motifs. This DNA binding function leads to the association of KAT6A with unmethylated CpG islands (CGIs) genome-wide. Mutation of the essential amino acids for DNA binding completely abrogates the enrichment of KAT6A at CGIs. In contrast, deletion of a second WH domain or the histone tail binding PHD fingers only subtly influences the binding of KAT6A to CGIs. Overexpression of a KAT6A WH1 mutant has a dominant negative effect on H3K9 histone acetylation, which is comparable to the effects upon overexpression of a KAT6A HAT domain mutant. Taken together, our work revealed a previously unrecognized chromatin recruitment mechanism of KAT6A, offering a new perspective on the role of KAT6A in gene regulation and human diseases.