Summary information and primary citation
- PDB-id
- 7y9y; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- RNA binding protein-RNA
- Method
- cryo-EM (2.77 Å)
- Summary
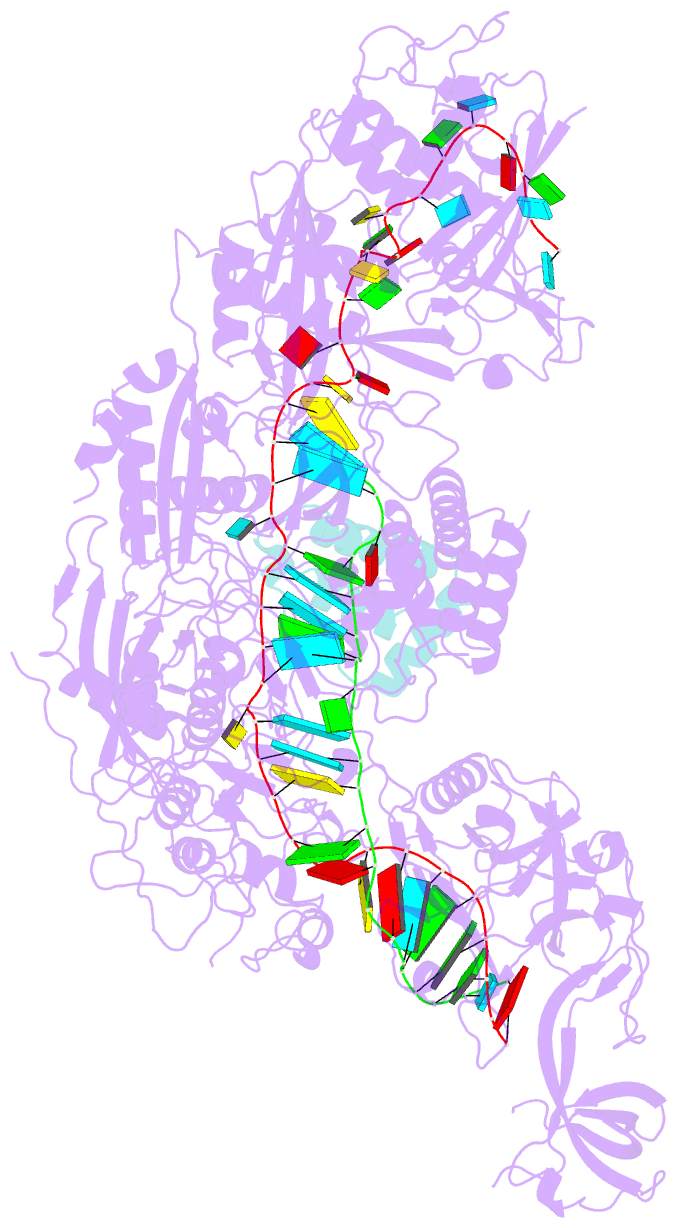
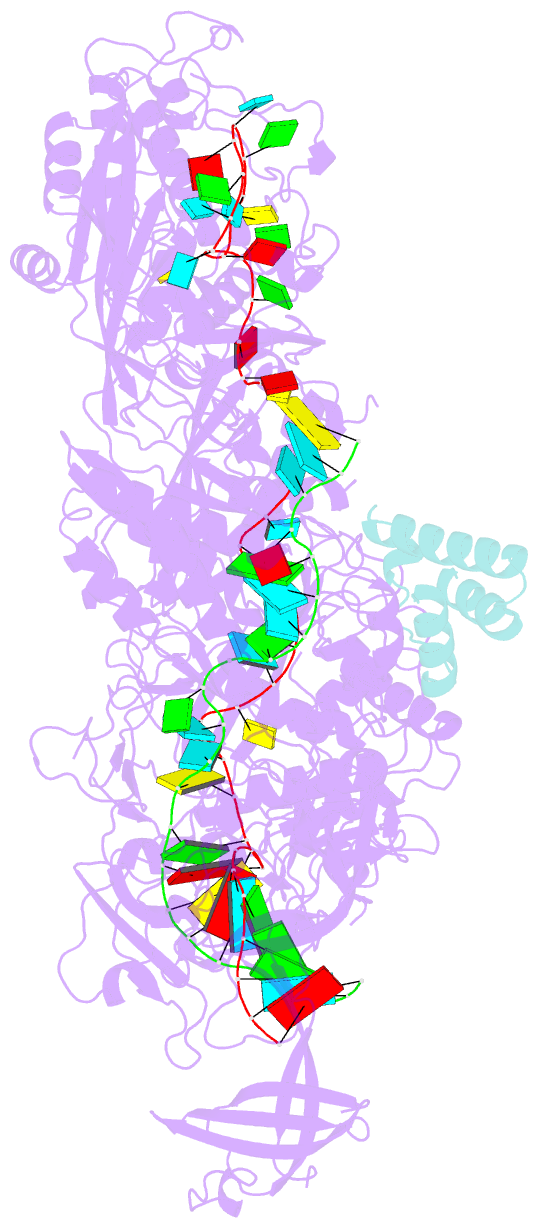
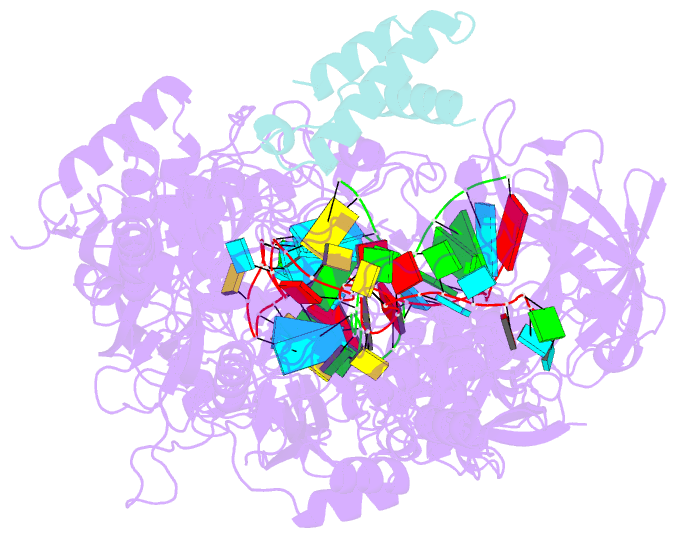
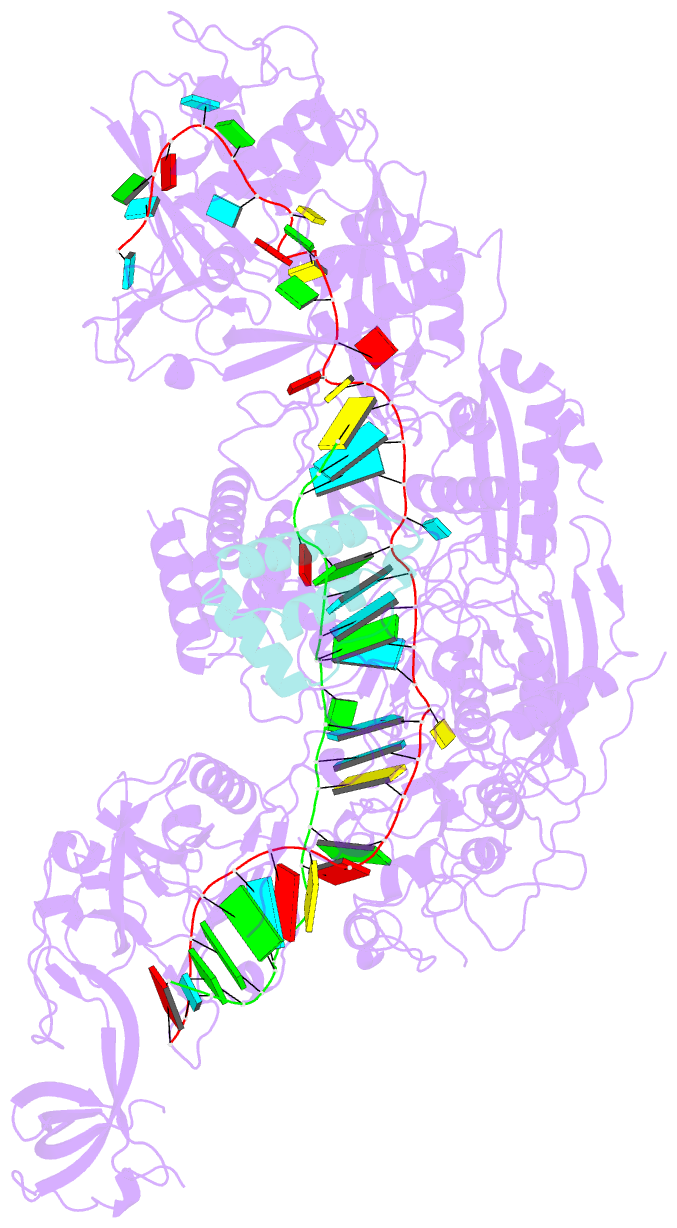
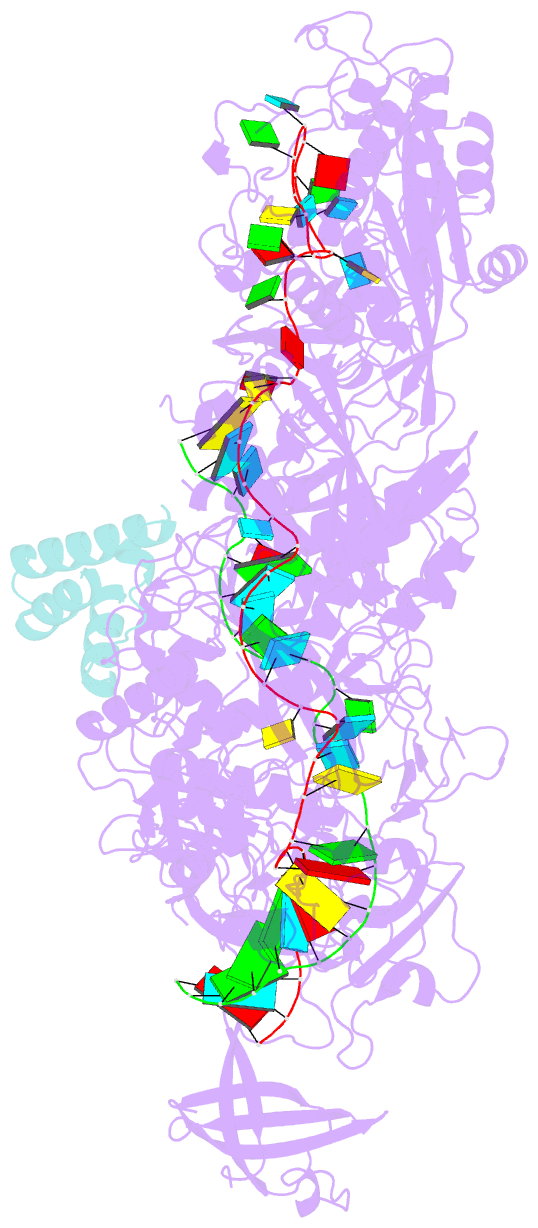
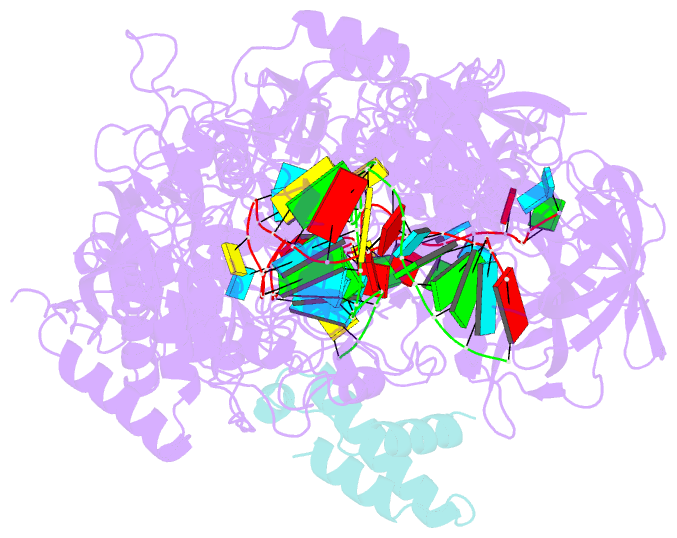
- Structure of the cas7-11-csx29-guide RNA-target RNA (no pfs) complex
- Reference
- Kato K, Okazaki S, Schmitt-Ulms C, Jiang K, Zhou W, Ishikawa J, Isayama Y, Adachi S, Nishizawa T, Makarova KS, Koonin EV, Abudayyeh OO, Gootenberg JS, Nishimasu H (2022): "RNA-triggered protein cleavage and cell growth arrest by the type III-E CRISPR nuclease-protease." Science, 378, 882-889. doi: 10.1126/science.add7347.
- Abstract
- The type III-E CRISPR-Cas7-11 effector binds a CRISPR RNA (crRNA) and the putative protease Csx29 and catalyzes crRNA-guided RNA cleavage. We report cryo-electron microscopy structures of the Cas7-11-crRNA-Csx29 complex with and without target RNA (tgRNA), and demonstrate that tgRNA binding induces conformational changes in Csx29. Biochemical experiments revealed tgRNA-dependent cleavage of the accessory protein Csx30 by Csx29. Reconstitution of the system in bacteria showed that Csx30 cleavage yields toxic protein fragments that cause growth arrest, which is regulated by Csx31. Csx30 binds Csx31 and the associated sigma factor RpoE (RNA polymerase, extracytoplasmic E), suggesting that Csx30-mediated RpoE inhibition modulates the cellular response to infection. We engineered the Cas7-11-Csx29-Csx30 system for programmable RNA sensing in mammalian cells. Overall, the Cas7-11-Csx29 effector is an RNA-dependent nuclease-protease.