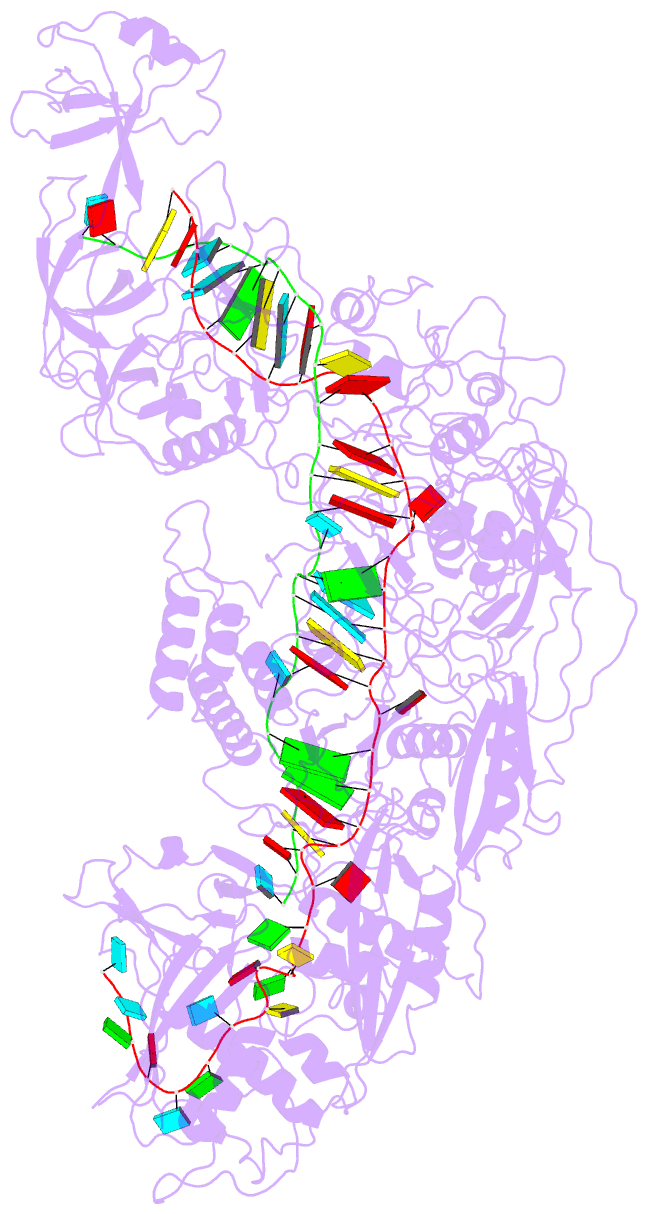
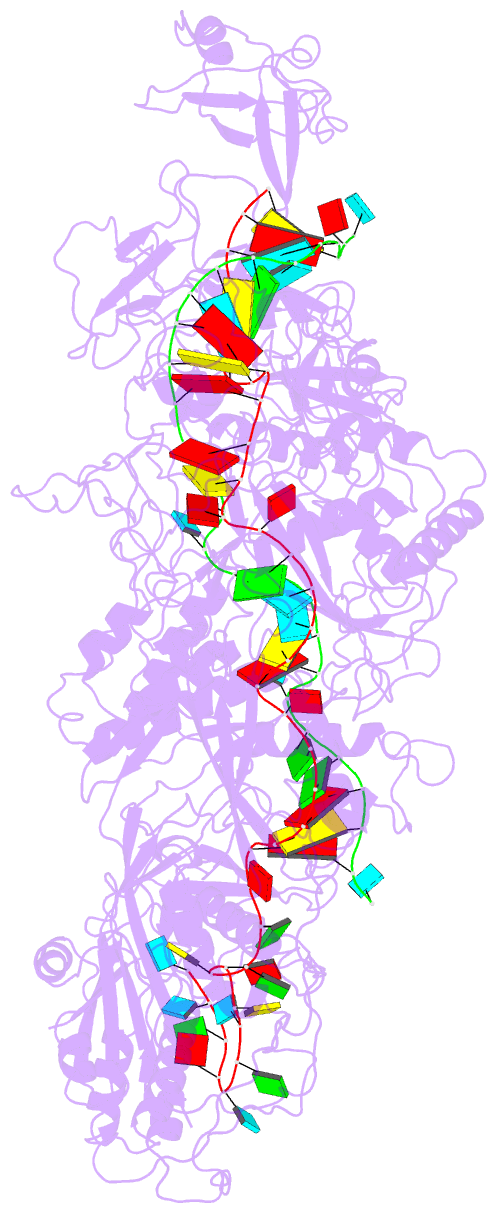
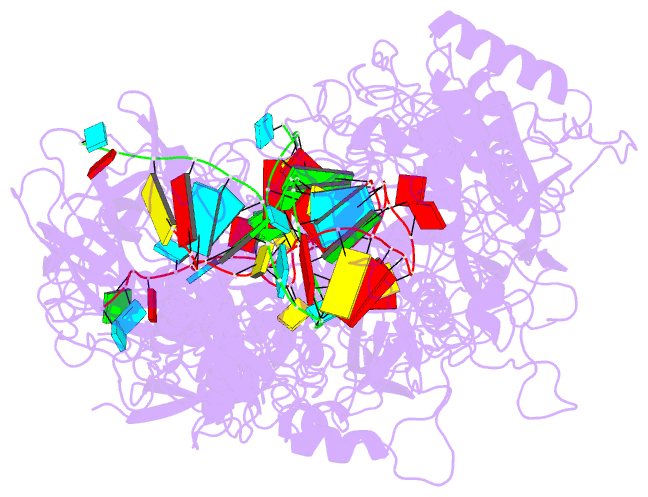
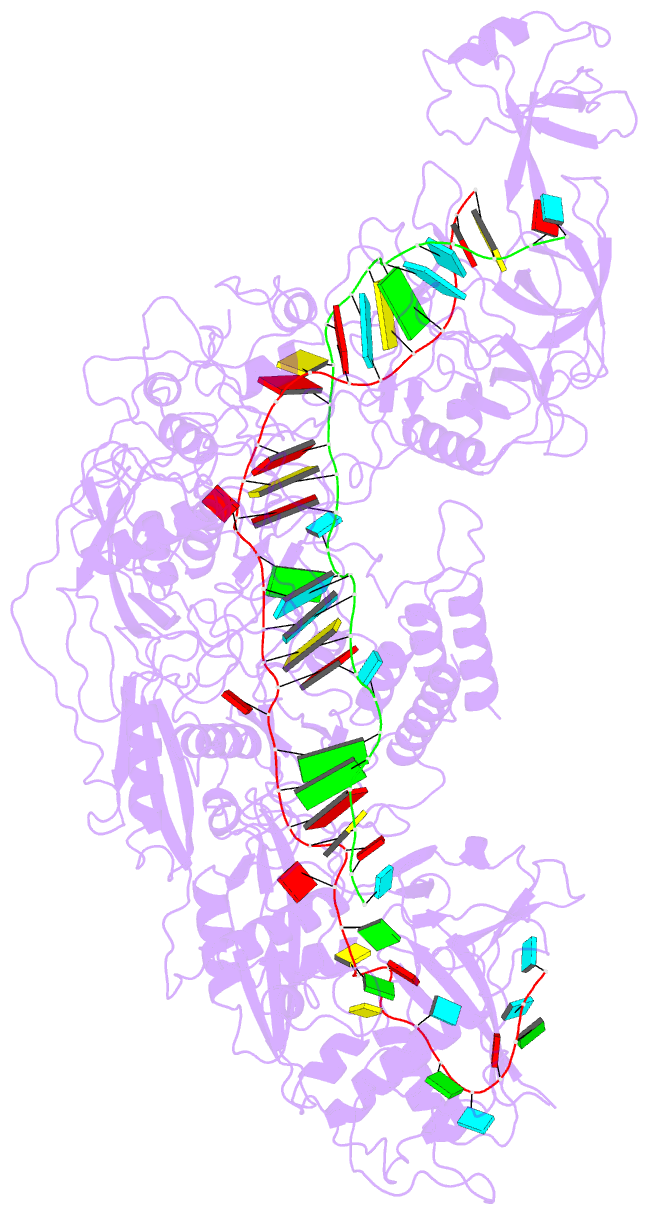
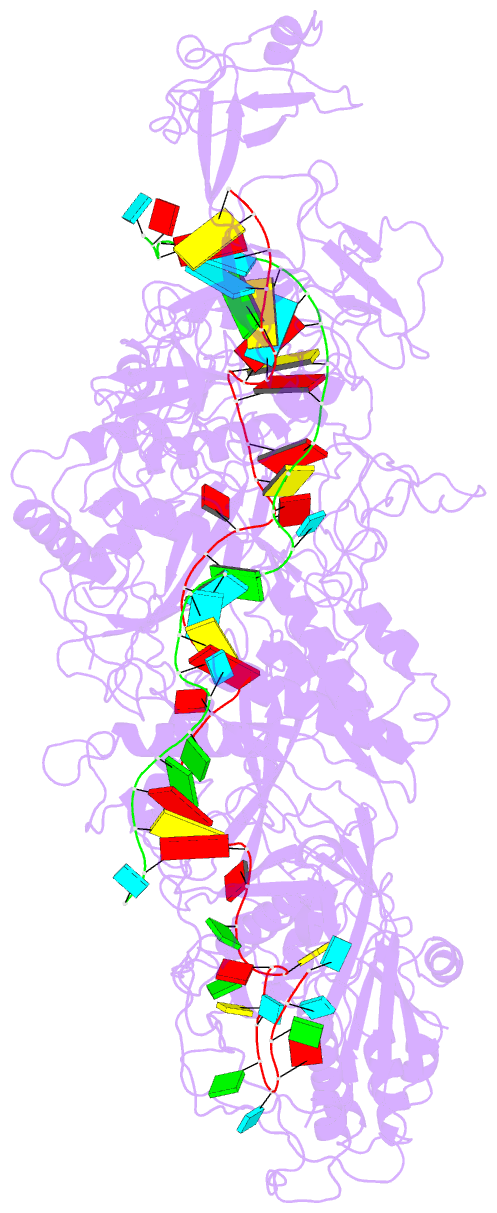
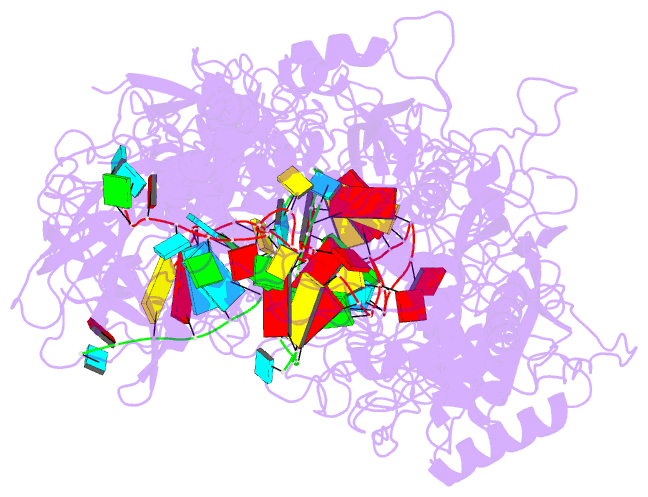
Summary information and primary citation
- PDB-id
- 7ynb; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- RNA binding protein
- Method
- cryo-EM (3.46 Å)
- Summary
- cryo-EM structure of cas7-11-crrna bound to target RNA-2
- Reference
- Huo Y, Zhao H, Dong Q, Jiang T (2023): "Cryo-EM structure and protease activity of the type III-E CRISPR-Cas effector." Nat Microbiol, 8, 522-532. doi: 10.1038/s41564-022-01316-4.
- Abstract
- The recently discovered type III-E CRISPR-Cas effector Cas7-11 shows promise when used as an RNA manipulation tool, but its structure and the mechanisms underlying its function remain unclear. Here we present four cryo-EM structures of Desulfonema ishimotonii Cas7-11-crRNA complex in pre-target and target RNA-bound states, and the cryo-EM structure of DiCas7-11-crRNA bound to its accessory protein DiCsx29. These data reveal structural elements for pre-crRNA processing, target RNA cleavage and regulation. Moreover, a 3' seed region of crRNA is involved in regulating RNA cleavage activity of DiCas7-11-crRNA-Csx29. Our analysis also shows that both the minimal mismatch of 4 nt to the 5' handle of crRNA and the minimal matching of the first 12 nt of the spacer by the target RNA are essential for triggering the protease activity of DiCas7-11-crRNA-Csx29 towards DiCsx30. Taken together, we propose that target RNA recognition and cleavage regulate and fine-tune the protease activity of DiCas7-11-crRNA-Csx29, thus preventing auto-immune responses.