Summary information and primary citation
- PDB-id
- 7ypq; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- DNA binding protein
- Method
- cryo-EM (3.1 Å)
- Summary
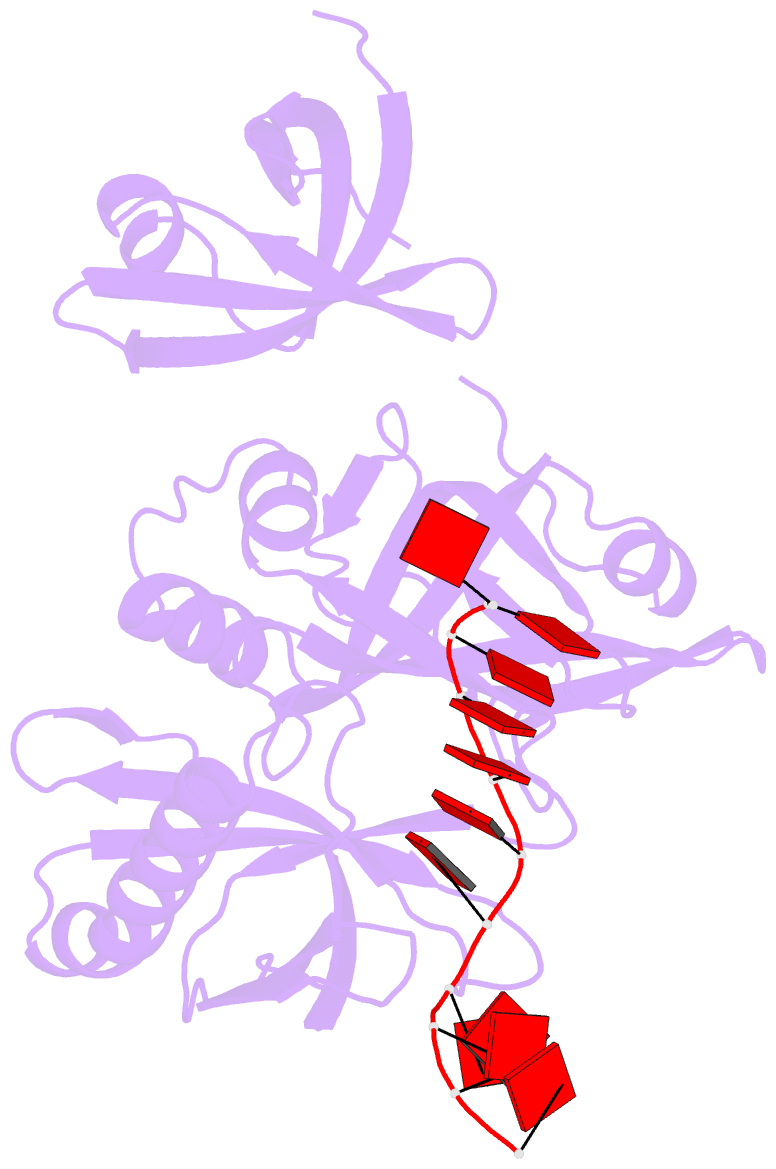
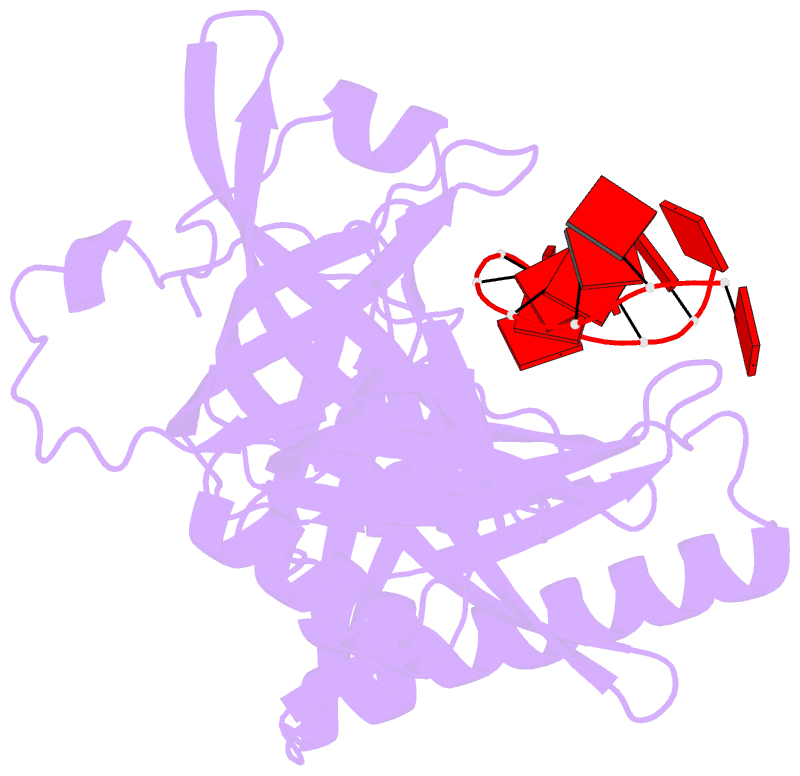
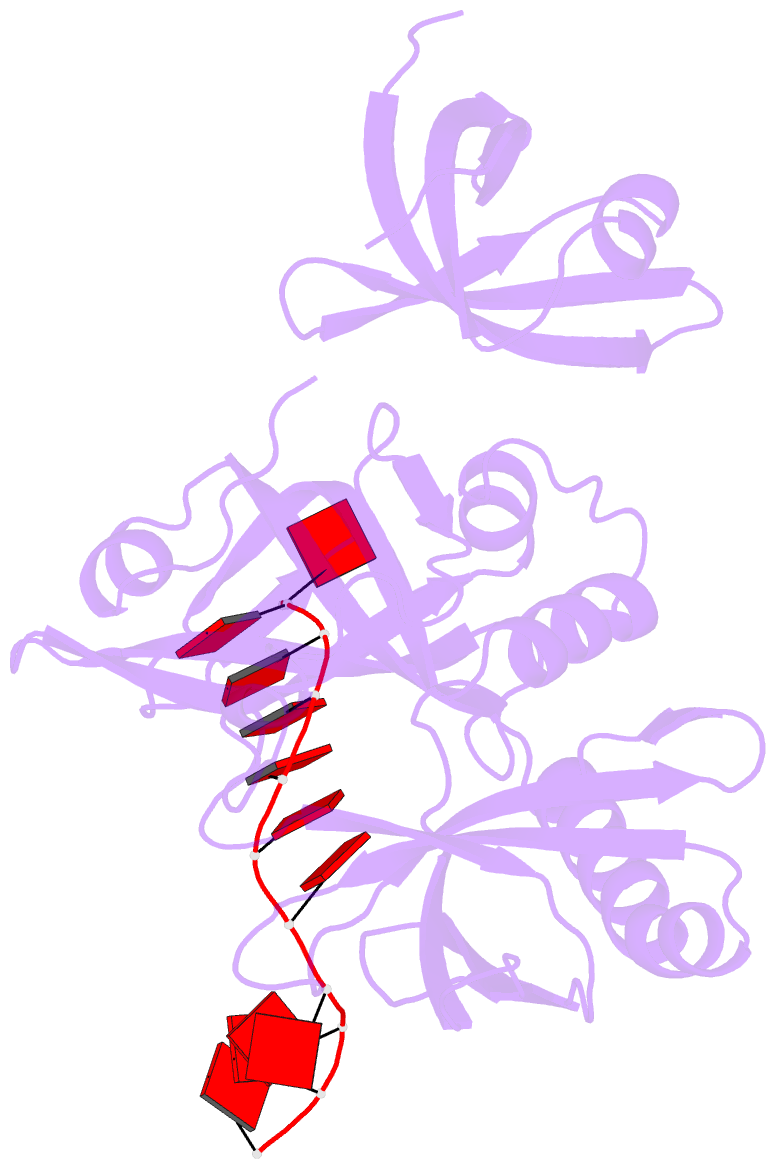
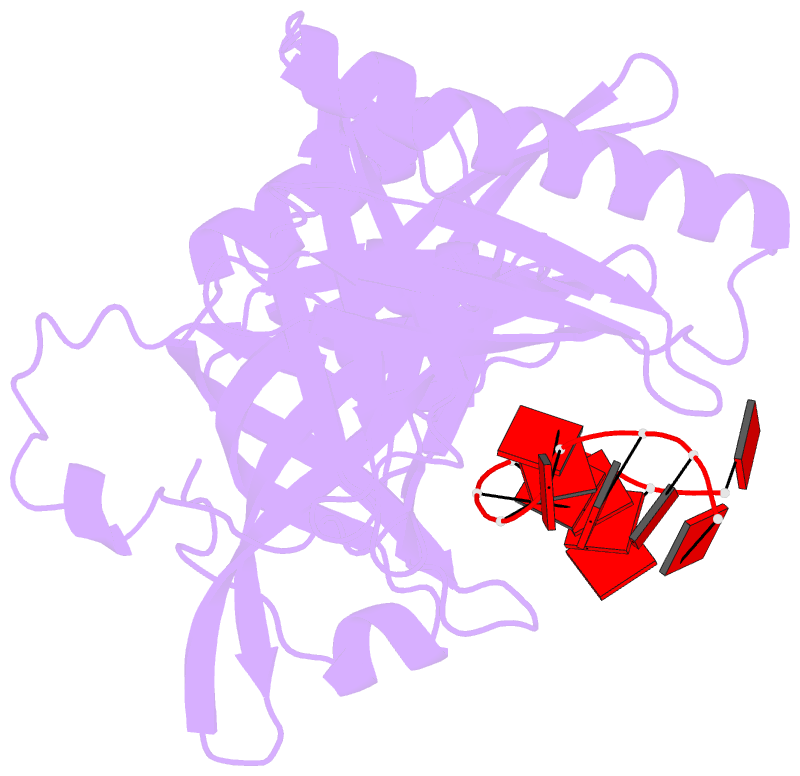
- cryo-EM structure of one baculovirus lef-3 molecule in complex with ssDNA
- Reference
- Yin J, Fu Y, Rao G, Li Z, Tian K, Chong T, Kuang K, Wang M, Hu Z, Cao S (2022): "Structural transitions during the cooperative assembly of baculovirus single-stranded DNA-binding protein on ssDNA." Nucleic Acids Res., 50, 13100-13113. doi: 10.1093/nar/gkac1142.
- Abstract
- Single-stranded DNA-binding proteins (SSBs) interact with single-stranded DNA (ssDNA) to form filamentous structures with various degrees of cooperativity, as a result of intermolecular interactions between neighboring SSB subunits on ssDNA. However, it is still challenging to perform structural studies on SSB-ssDNA filaments at high resolution using the most studied SSB models, largely due to the intrinsic flexibility of these nucleoprotein complexes. In this study, HaLEF-3, an SSB protein from Helicoverpa armigera nucleopolyhedrovirus, was used for in vitro assembly of SSB-ssDNA filaments, which were structurally studied at atomic resolution using cryo-electron microscopy. Combined with the crystal structure of ssDNA-free HaLEF-3 octamers, our results revealed that the three-dimensional rearrangement of HaLEF-3 induced by an internal hinge-bending movement is essential for the formation of helical SSB-ssDNA complexes, while the contacting interface between adjacent HaLEF-3 subunits remains basically intact. We proposed a local cooperative SSB-ssDNA binding model, in which, triggered by exposure to oligonucleotides, HaLEF-3 molecules undergo ring-to-helix transition to initiate continuous SSB-SSB interactions along ssDNA. Unique structural features revealed by the assembly of HaLEF-3 on ssDNA suggest that HaLEF-3 may represent a new class of SSB.