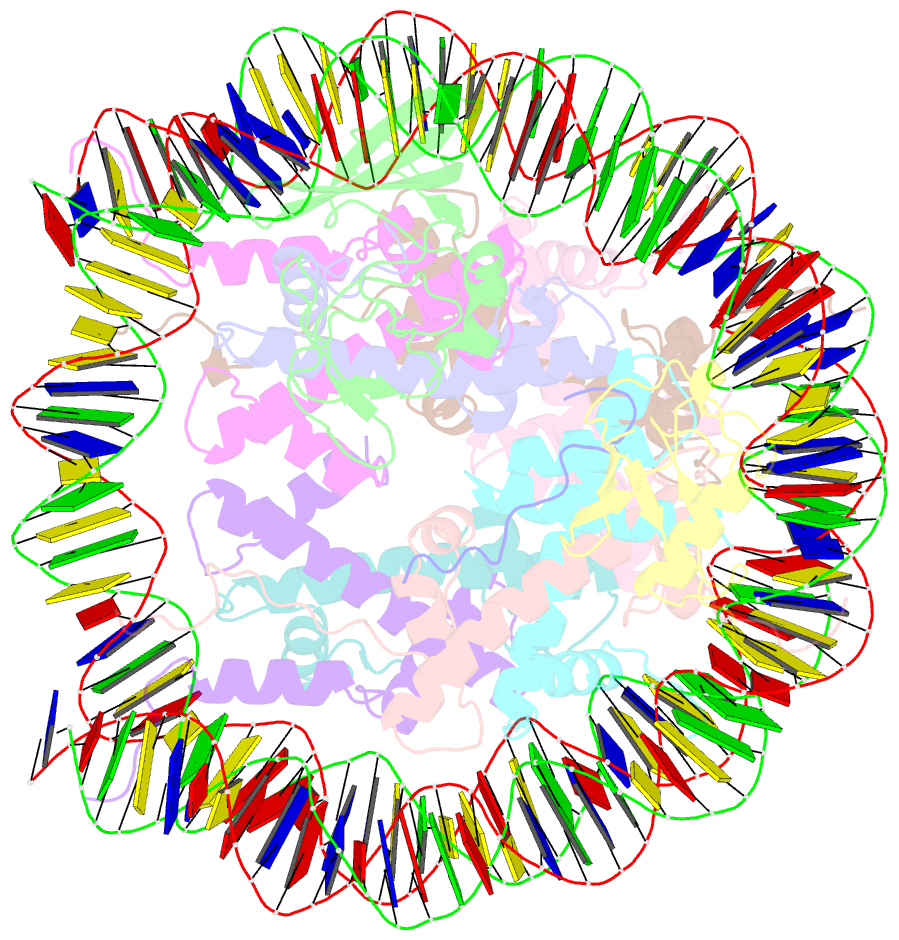
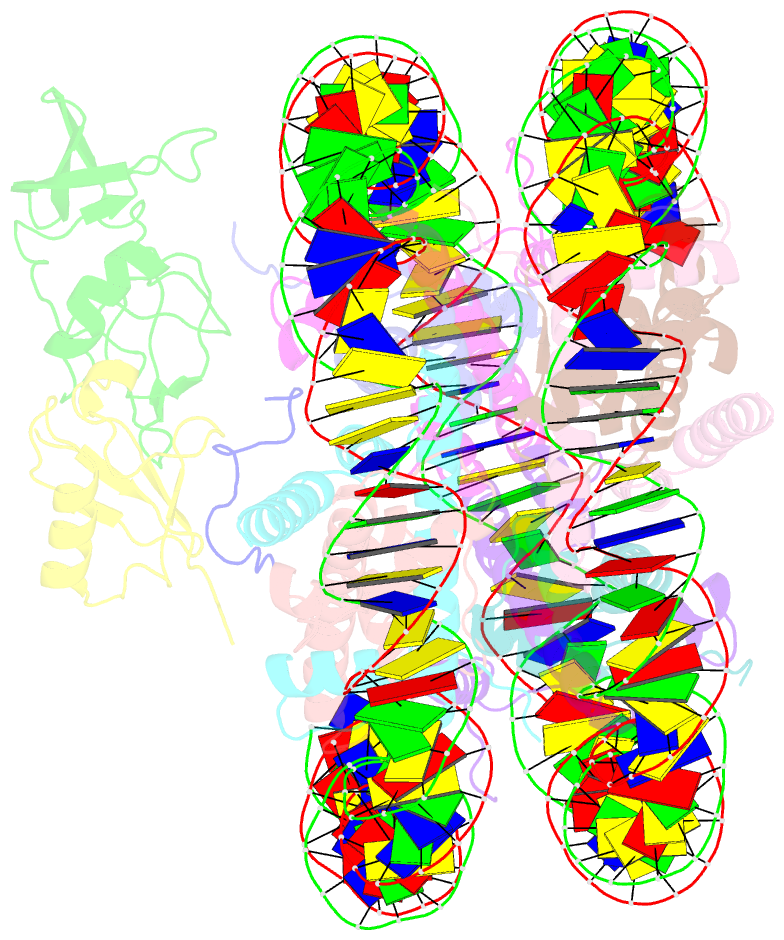
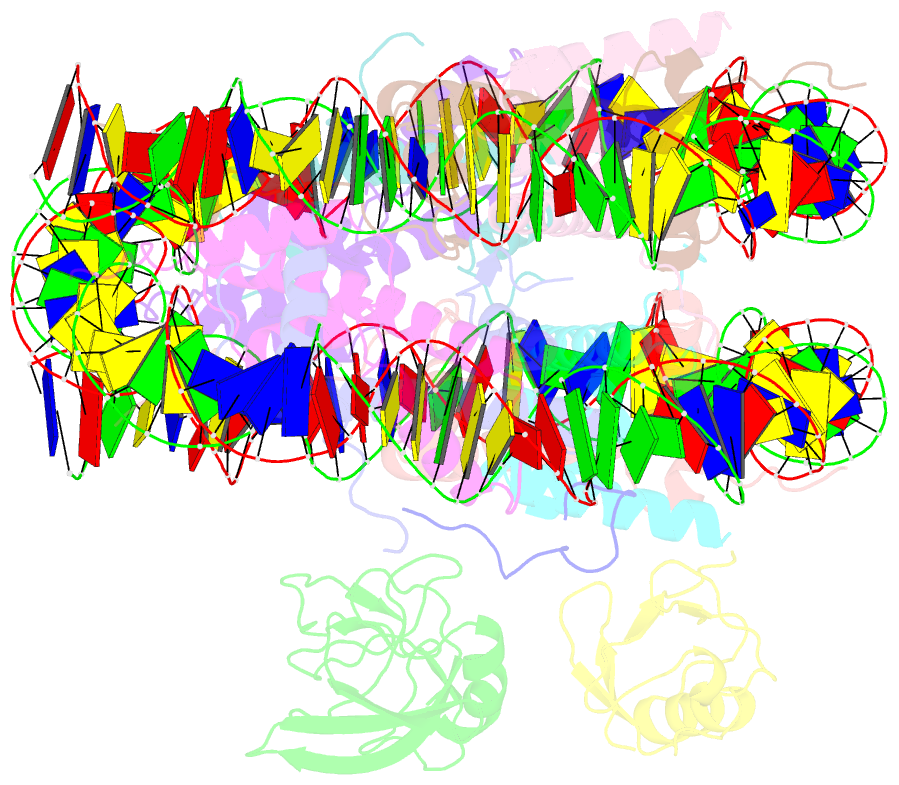
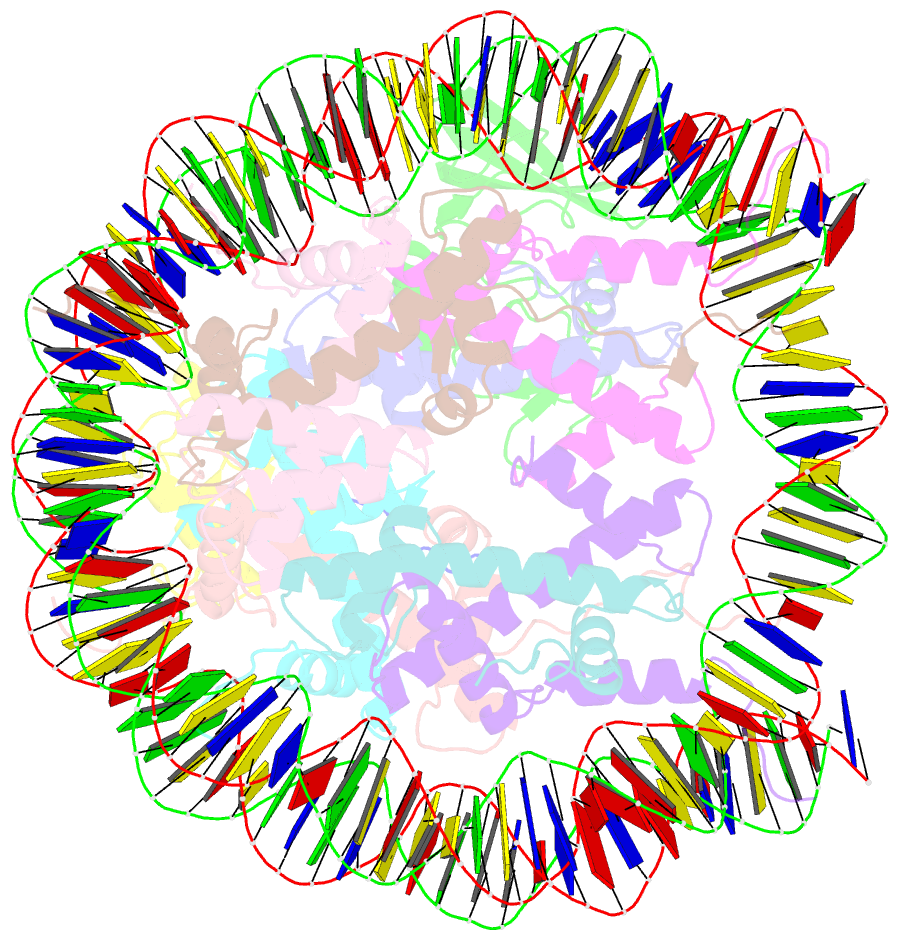
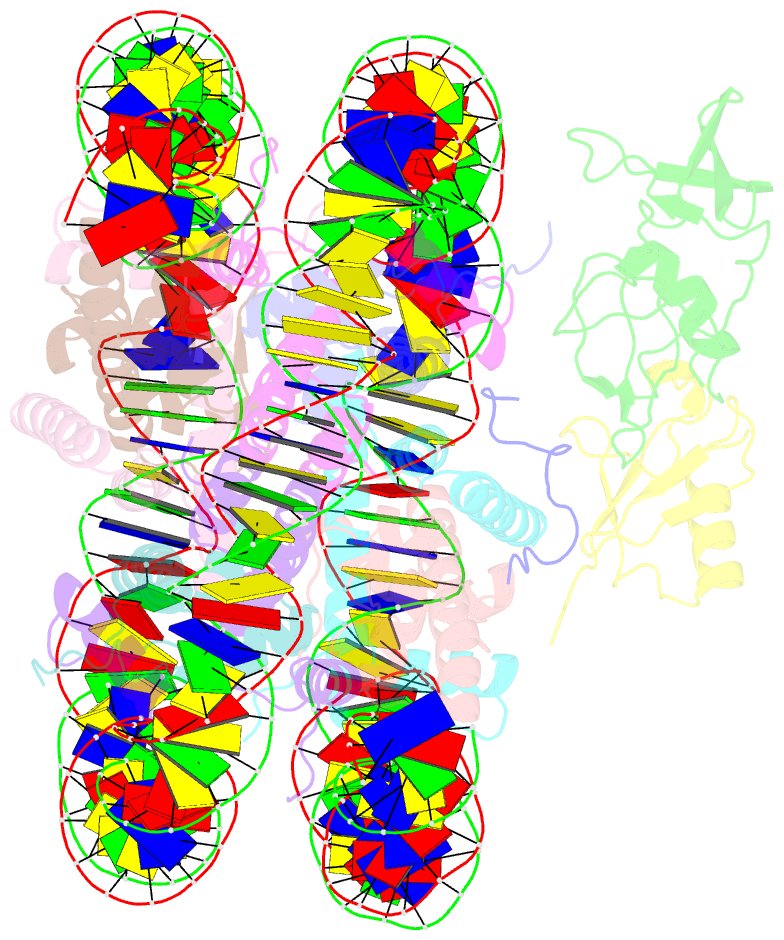
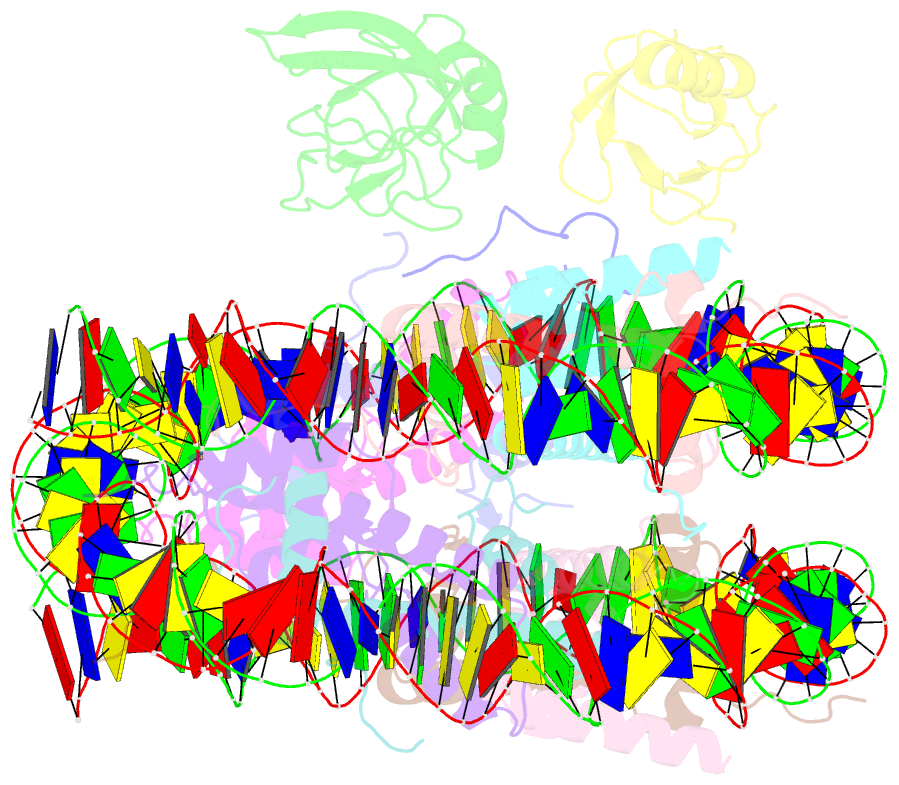
Summary information and primary citation
- PDB-id
- 7yqk; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- nuclear protein
- Method
- cryo-EM (3.38 Å)
- Summary
- cryo-EM structure of gammah2axk15ub-h4k20me2 nucleosome bound to 53bp1
- Reference
- Ai H, Chu GC, Gong Q, Tong ZB, Deng Z, Liu X, Yang F, Xu Z, Li JB, Tian C, Liu L (2022): "Chemical Synthesis of Post-Translationally Modified H2AX Reveals Redundancy in Interplay between Histone Phosphorylation, Ubiquitination, and Methylation on the Binding of 53BP1 with Nucleosomes." J.Am.Chem.Soc., 144, 18329-18337. doi: 10.1021/jacs.2c06156.
- Abstract
- The chemical synthesis of homogeneously modified histones is a powerful approach to quantitatively decipher how post-translational modifications (PTMs) modulate epigenetic events. Herein, we describe the expedient syntheses of a selection of phosphorylated and ubiquitinated H2AX proteins in a strategy integrating expressed protein hydrazinolysis and auxiliary-mediated protein ligation. These modified H2AX proteins were then used to discover that although H2AXS139 phosphorylation can enhance the binding of the DNA damage repair factor 53BP1 to either an unmodified nucleosome or that bearing a single H2AXK15ub or H4K20me2 modification, it augments 53BP1's binding only weakly to nucleosomes bearing both H2AXK15ub and H4K20me2. To better understand why such a trivalent additive effect is lacking, we solved the cryo-EM structure (3.38 Å) of the complex of 53BP1 with the H2AXK15ub/S139ph_H4K20me2 nucleosome, which showed that H2AXS139 phosphorylation distorts the interaction interface between ubiquitin and 53BP1's UDR motif. Our study revealed that there is redundancy in the interplay of multiple histone PTMs, which may be useful for controlling the dynamic distribution of effector proteins onto nucleosomes bearing different histone variants and PTMs in a time-dependent fashion, through specific cellular biochemical events.