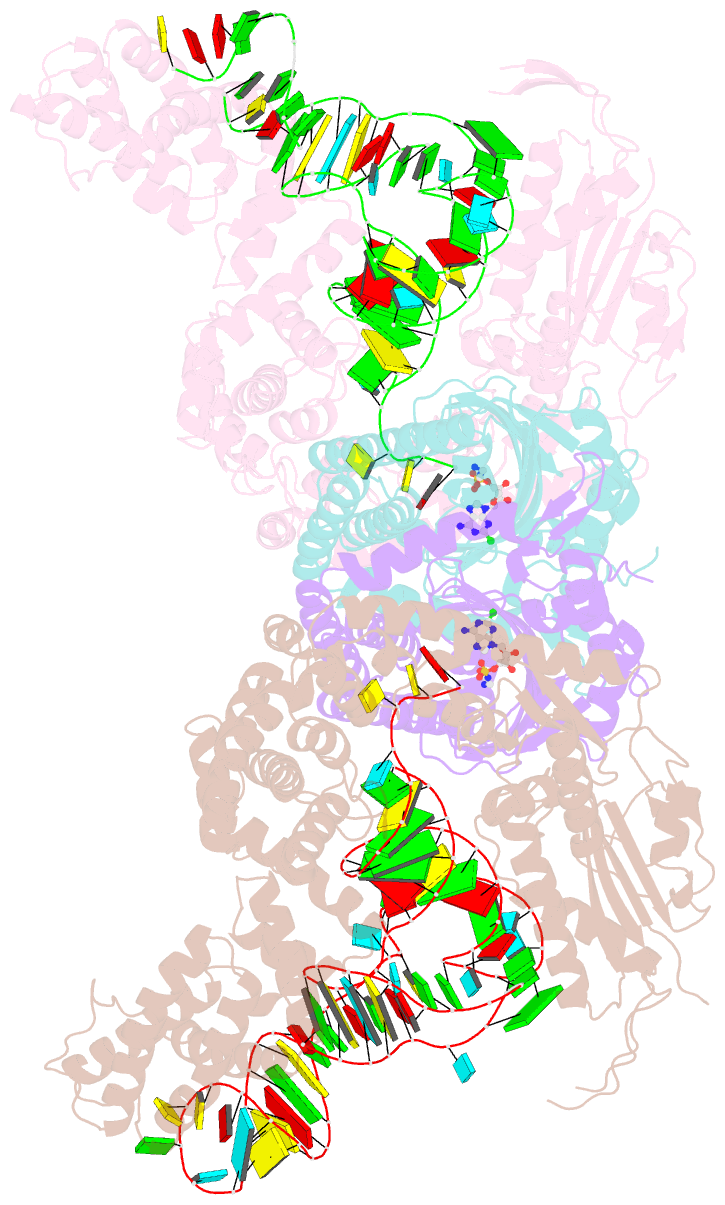
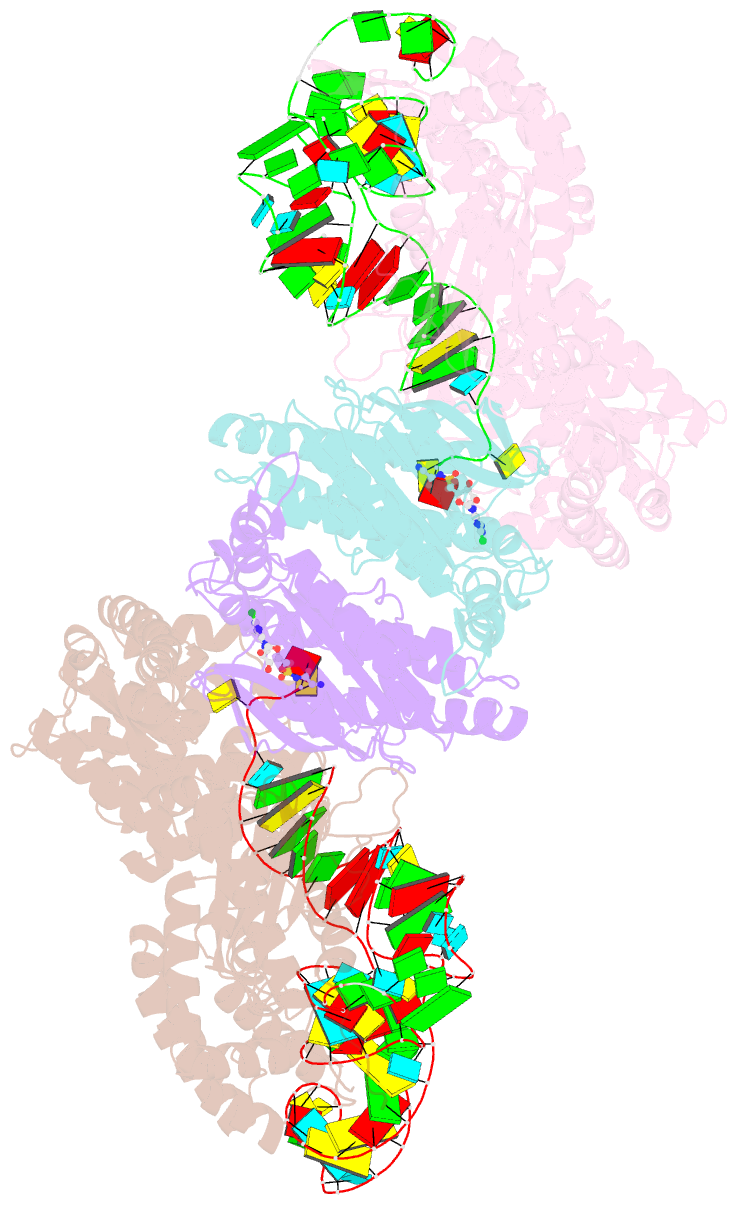
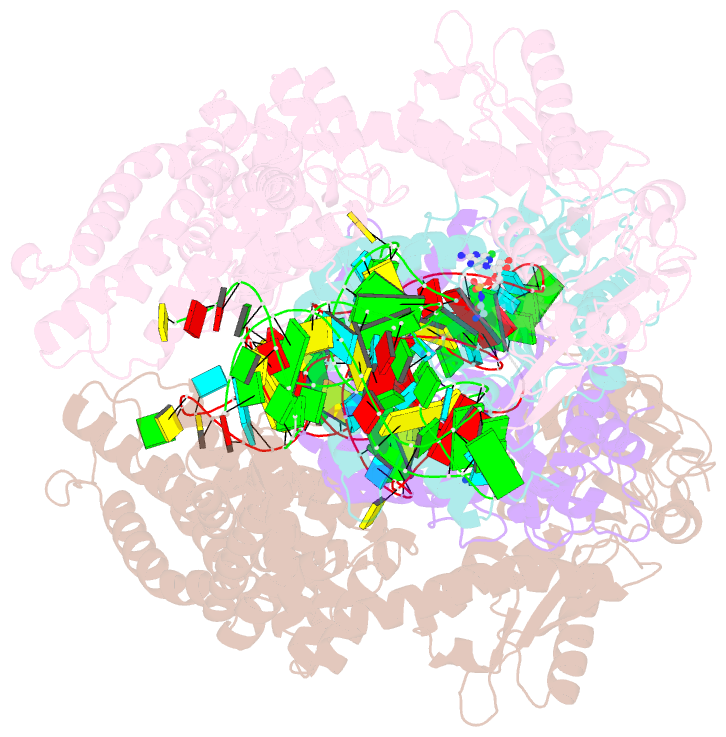
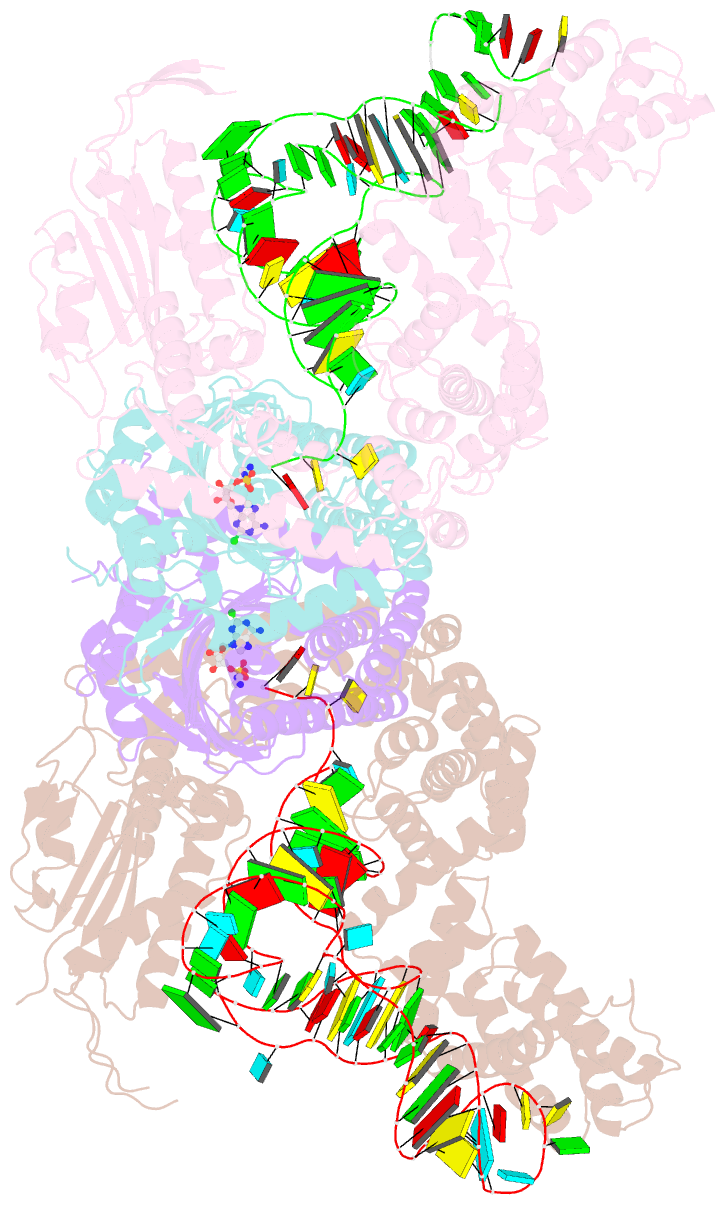
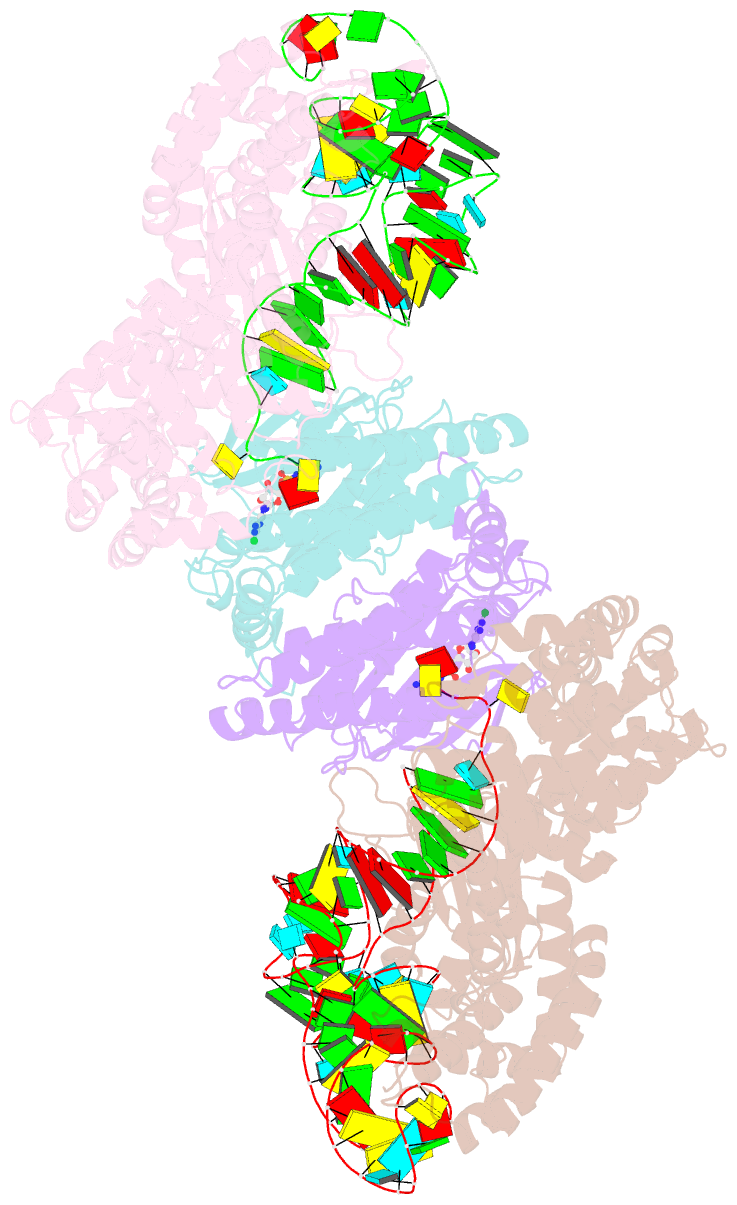
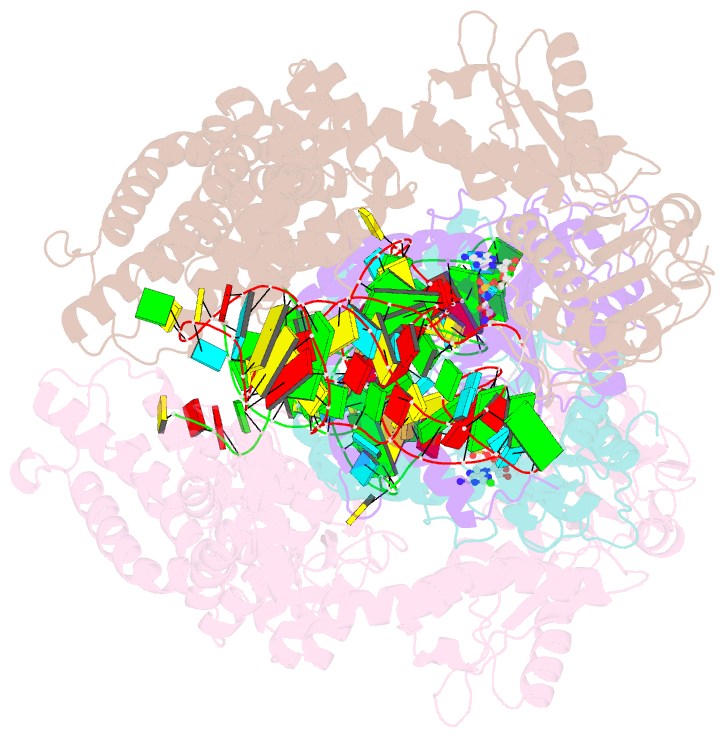
Summary information and primary citation
- PDB-id
- 7yse; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transferase-RNA
- Method
- X-ray (2.907 Å)
- Summary
- Crystal structure of e. coli heterotetrameric glyrs in complex with trna
- Reference
- Han L, Luo Z, Ju Y, Chen B, Zou T, Wang J, Xu J, Gu Q, Yang XL, Schimmel P, Zhou H (2023): "The binding mode of orphan glycyl-tRNA synthetase with tRNA supports the synthetase classification and reveals large domain movements." Sci Adv, 9, eadf1027. doi: 10.1126/sciadv.adf1027.
- Abstract
- As a class of essential enzymes in protein translation, aminoacyl-transfer RNA (tRNA) synthetases (aaRSs) are organized into two classes of 10 enzymes each, based on two conserved active site architectures. The (αβ)2 glycyl-tRNA synthetase (GlyRS) in many bacteria is an orphan aaRS whose sequence and unprecedented X-shaped structure are distinct from those of all other aaRSs, including many other bacterial and all eukaryotic GlyRSs. Here, we report a cocrystal structure to elucidate how the orphan GlyRS kingdom specifically recognizes its substrate tRNA. This structure is sharply different from those of other aaRS-tRNA complexes but conforms to the clash-free, cross-class aaRS-tRNA docking found with conventional structures and reinforces the class-reconstruction paradigm. In addition, noteworthy, the X shape of orphan GlyRS is condensed with the largest known spatial rearrangement needed by aaRSs to capture tRNAs, which suggests potential nonactive site targets for aaRS-directed antibiotics, instead of less differentiated hard-to-drug active site locations.