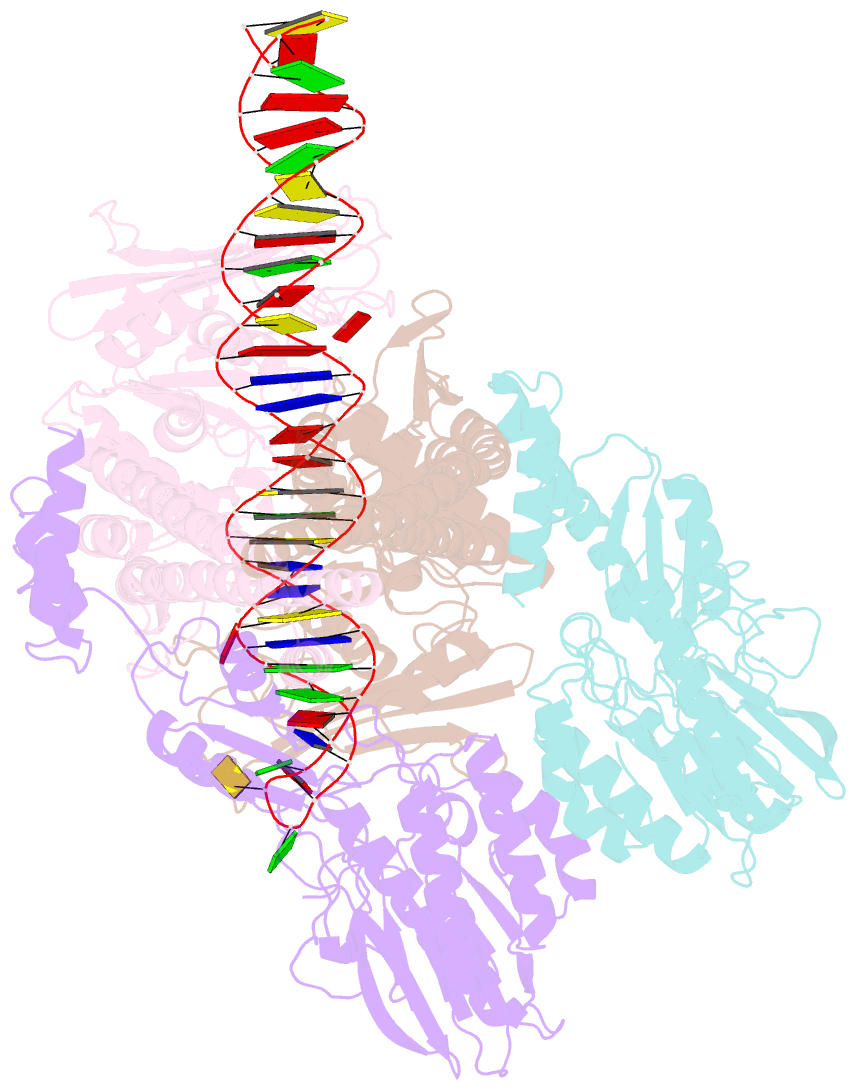
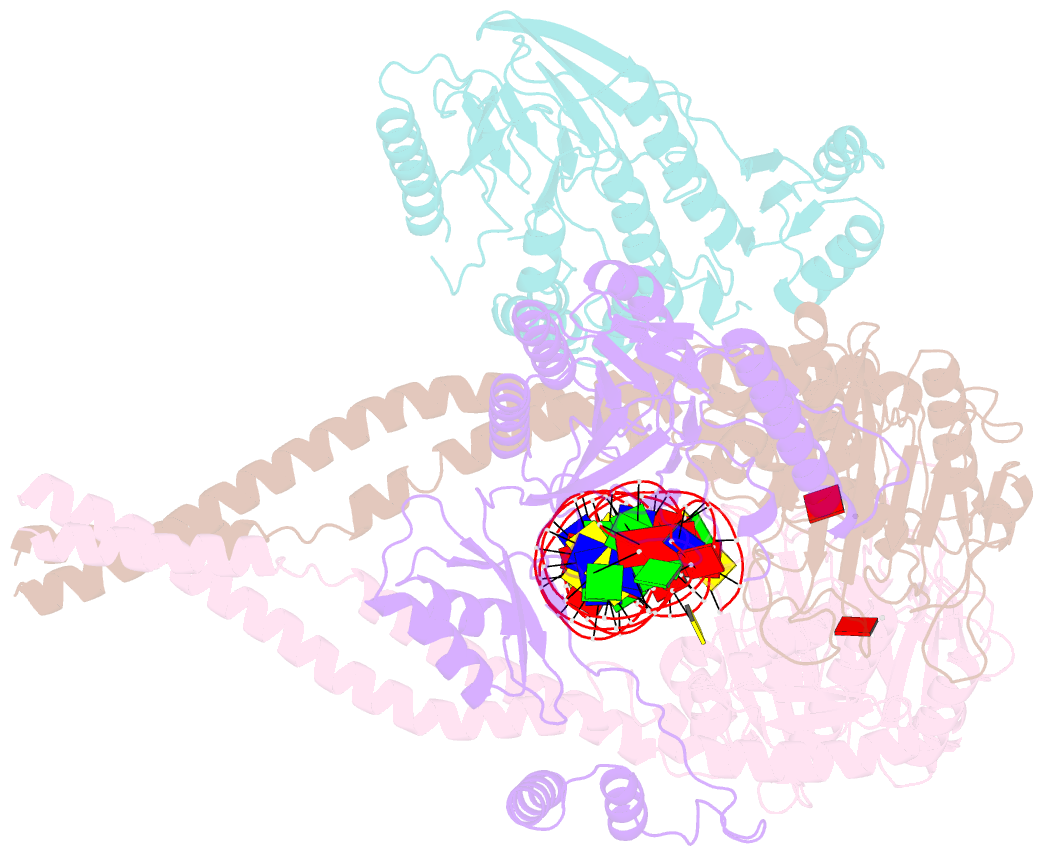
Summary information and primary citation
- PDB-id
- 7yzp; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- DNA binding protein
- Method
- cryo-EM (4.0 Å)
- Summary
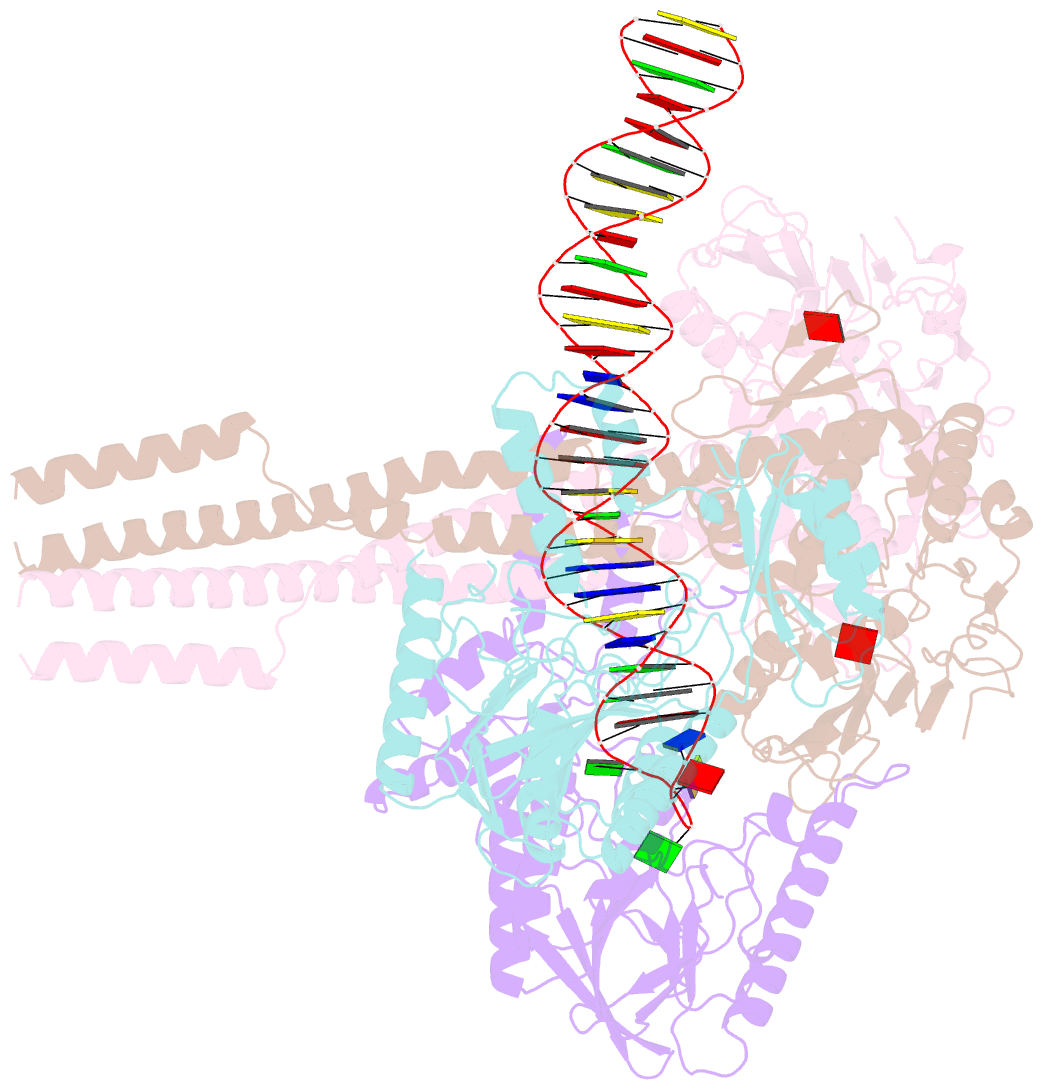
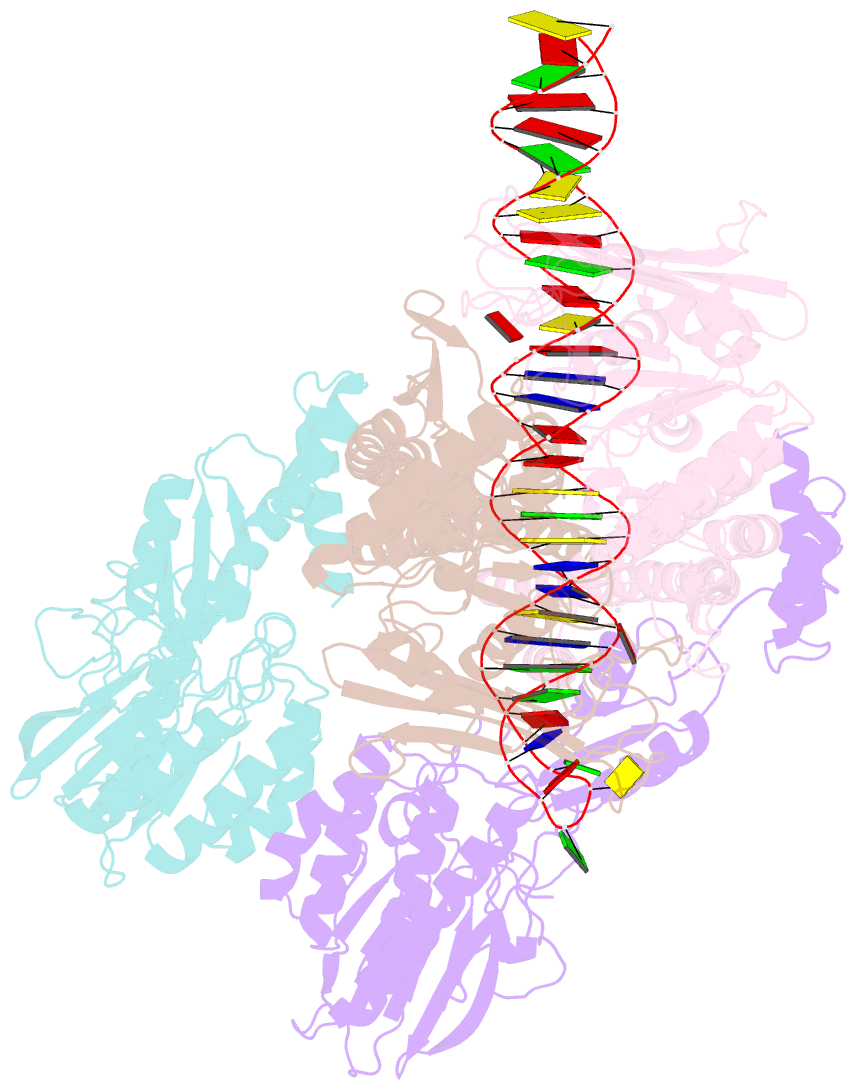
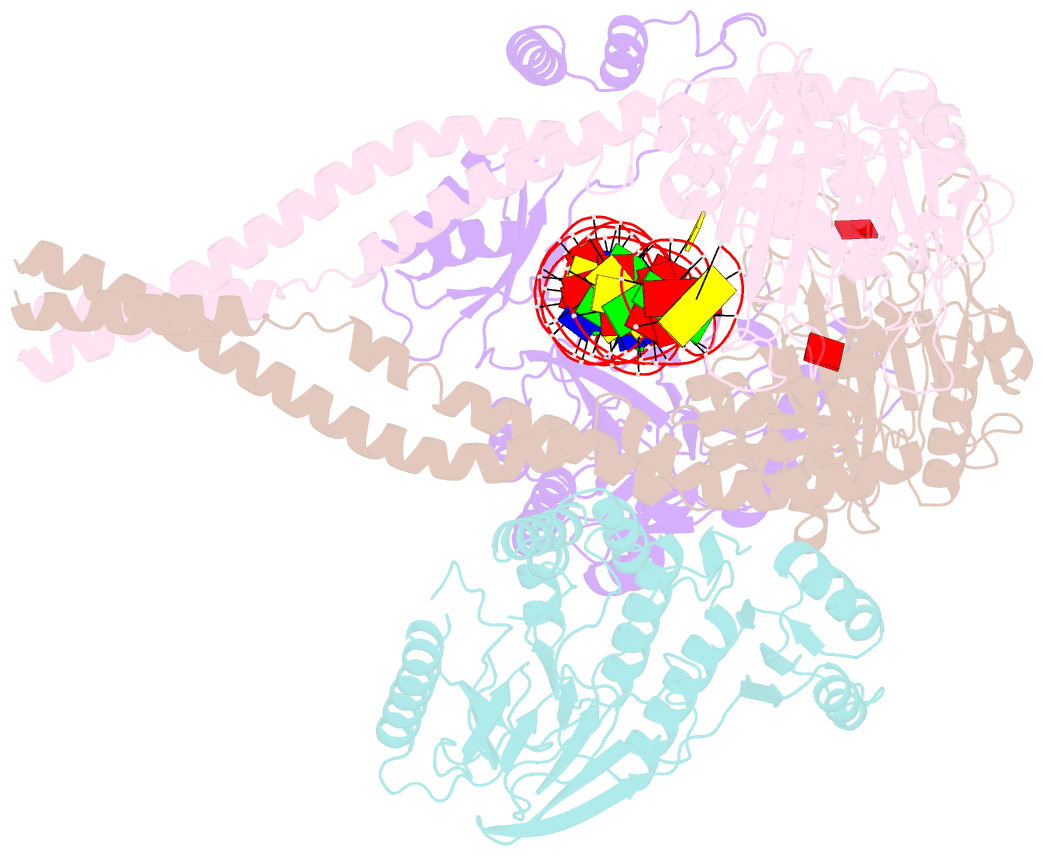
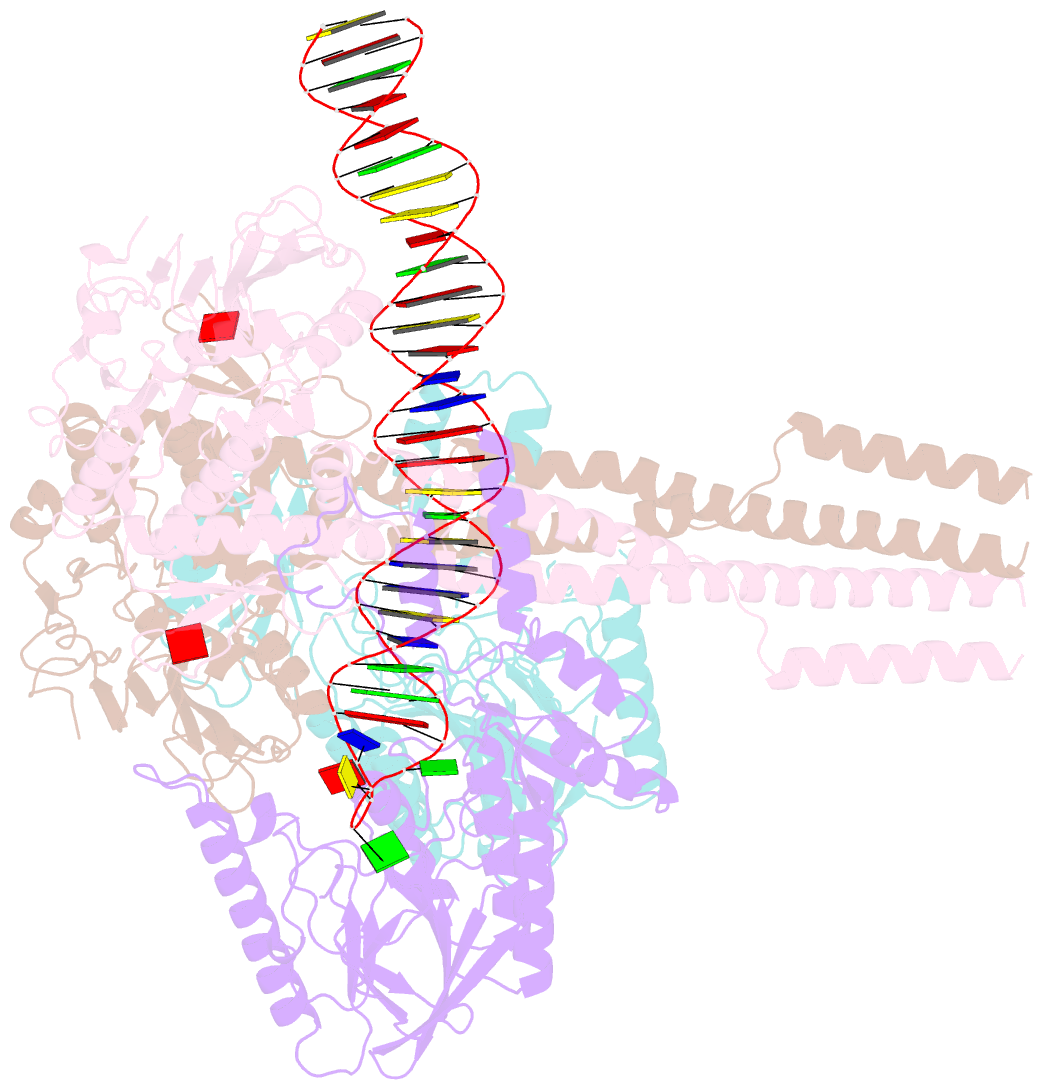
- Hairpin-bound state of the e. coli mre11-rad50 (sbccd) head complex bound to adp and a DNA hairpin
- Reference
- Gut F, Kashammer L, Lammens K, Bartho JD, Boggusch AM, van de Logt E, Kessler B, Hopfner KP (2022): "Structural mechanism of endonucleolytic processing of blocked DNA ends and hairpins by Mre11-Rad50." Mol.Cell, 82, 3513-3522.e6. doi: 10.1016/j.molcel.2022.07.019.
- Abstract
- DNA double-strand breaks (DSBs) threaten genome stability and are linked to tumorigenesis in humans. Repair of DSBs requires the removal of attached proteins and hairpins through a poorly understood but physiologically critical endonuclease activity by the Mre11-Rad50 complex. Here, we report cryoelectron microscopy (cryo-EM) structures of the bacterial Mre11-Rad50 homolog SbcCD bound to a protein-blocked DNA end and a DNA hairpin. The structures reveal that Mre11-Rad50 bends internal DNA for endonucleolytic cleavage and show how internal DNA, DNA ends, and hairpins are processed through a similar ATP-regulated conformational state. Furthermore, Mre11-Rad50 is loaded onto blocked DNA ends with Mre11 pointing away from the block, explaining the distinct biochemistries of 3' → 5' exonucleolytic and endonucleolytic incision through the way Mre11-Rad50 interacts with diverse DNA ends. In summary, our results unify Mre11-Rad50's enigmatic nuclease diversity within a single structural framework and reveal how blocked DNA ends and hairpins are processed.