Summary information and primary citation
- PDB-id
- 7z87; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- DNA binding protein
- Method
- cryo-EM (2.91 Å)
- Summary
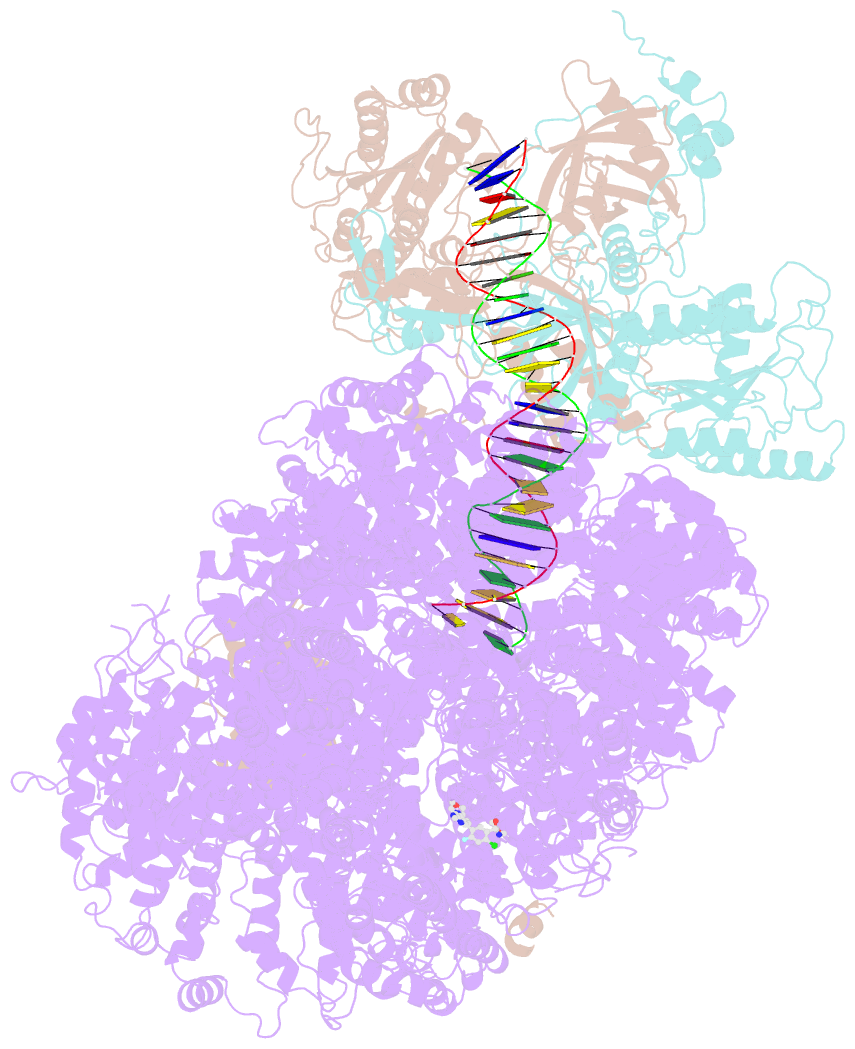
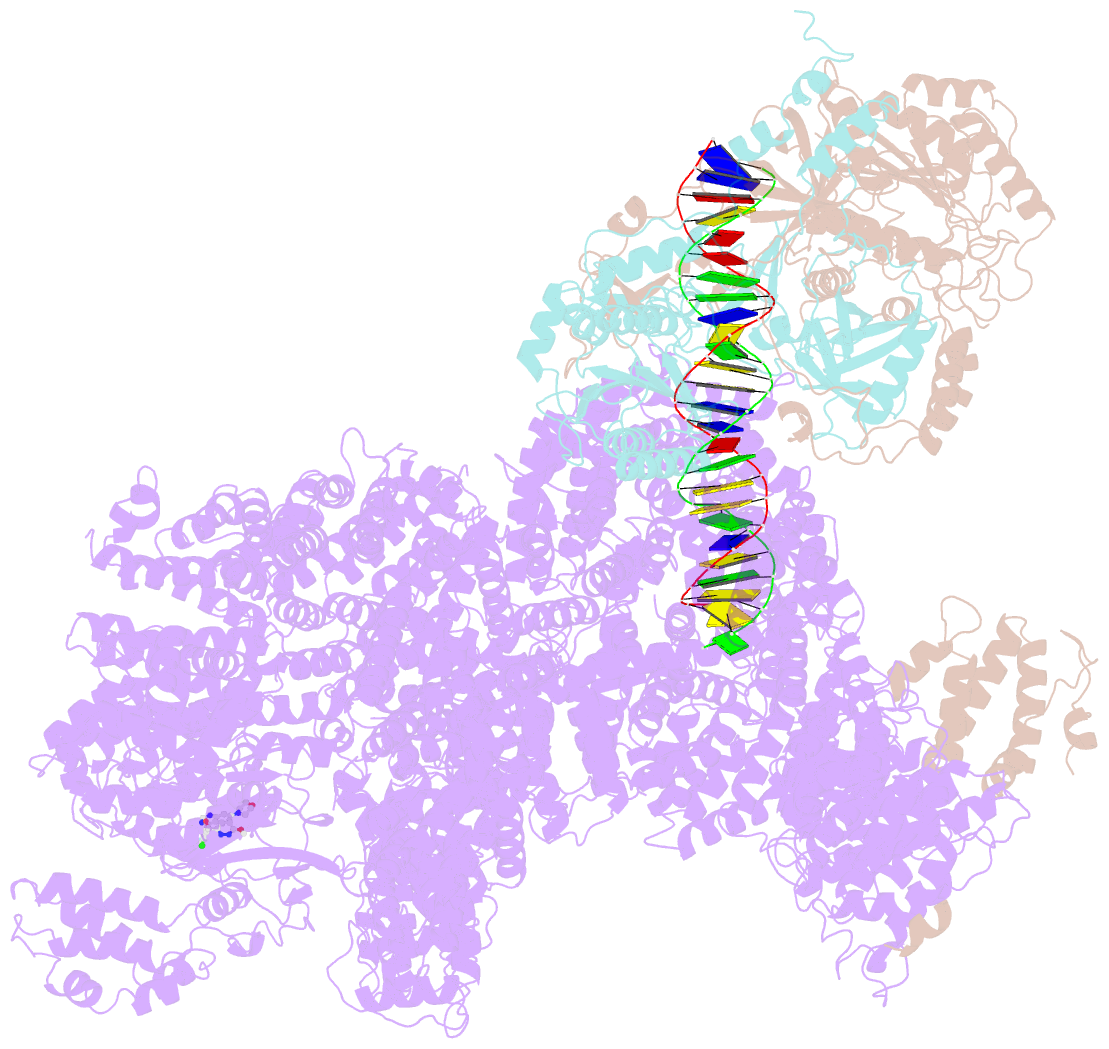
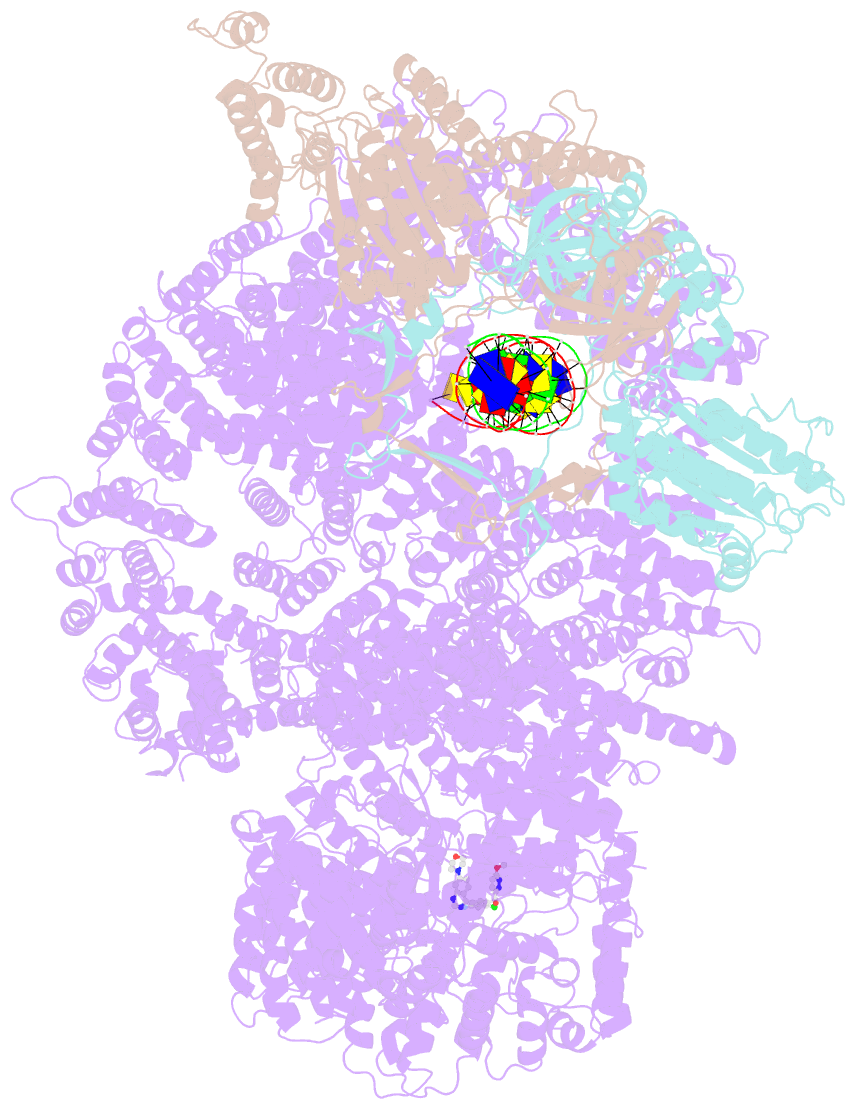
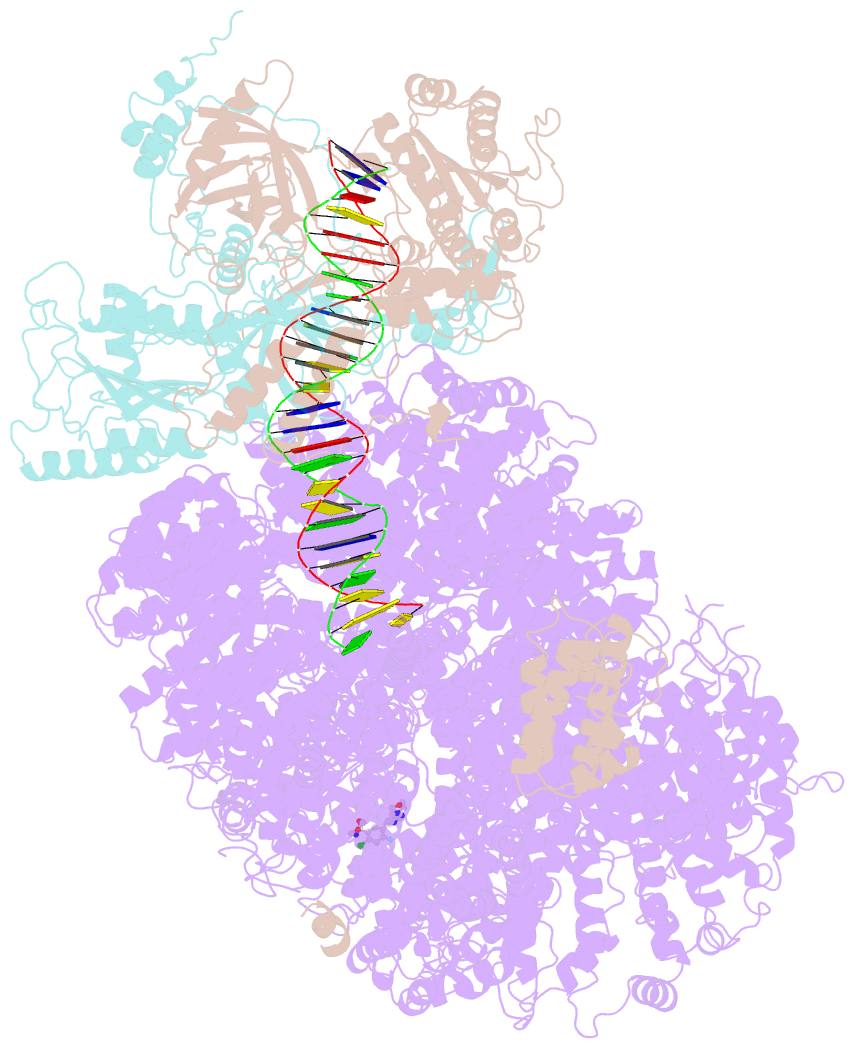
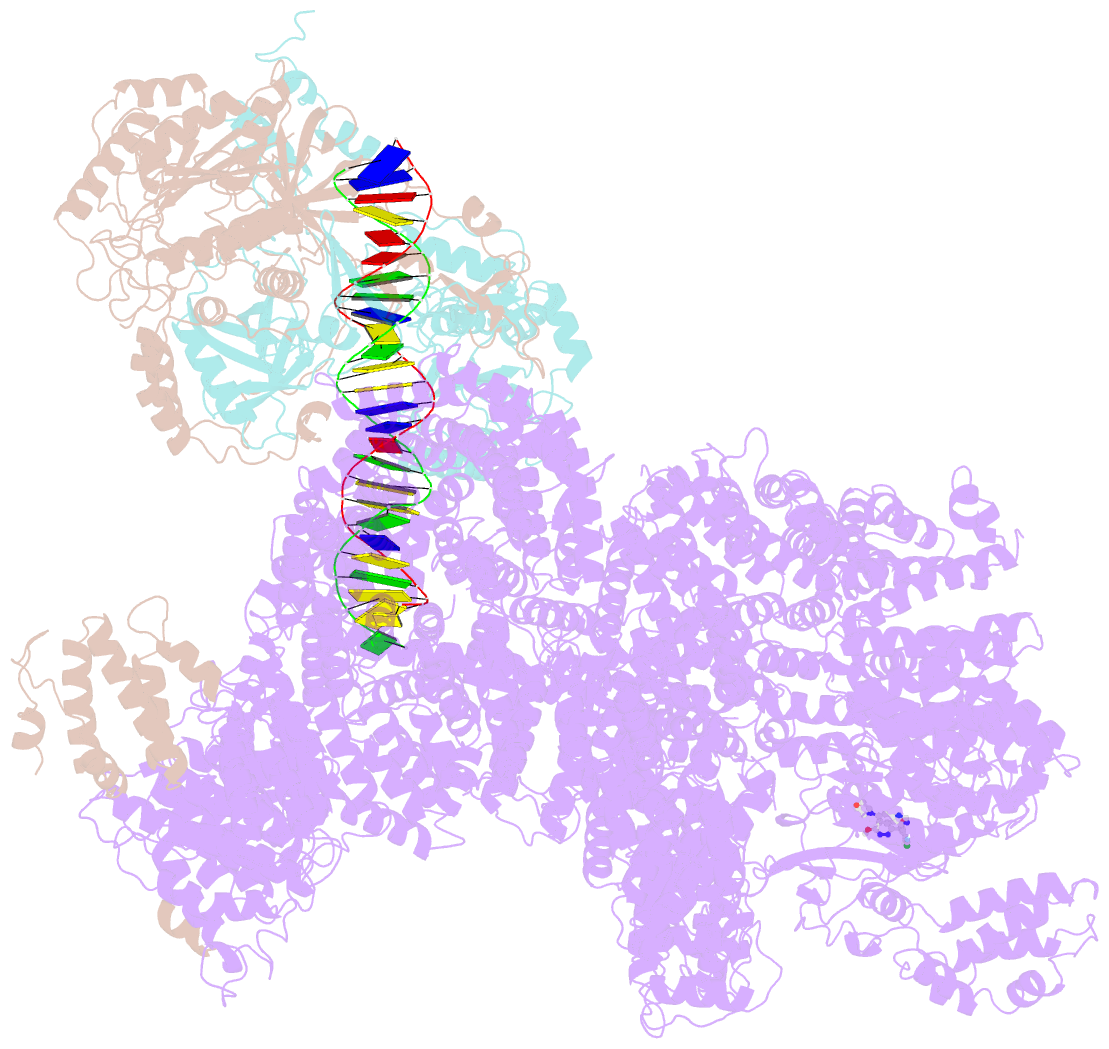
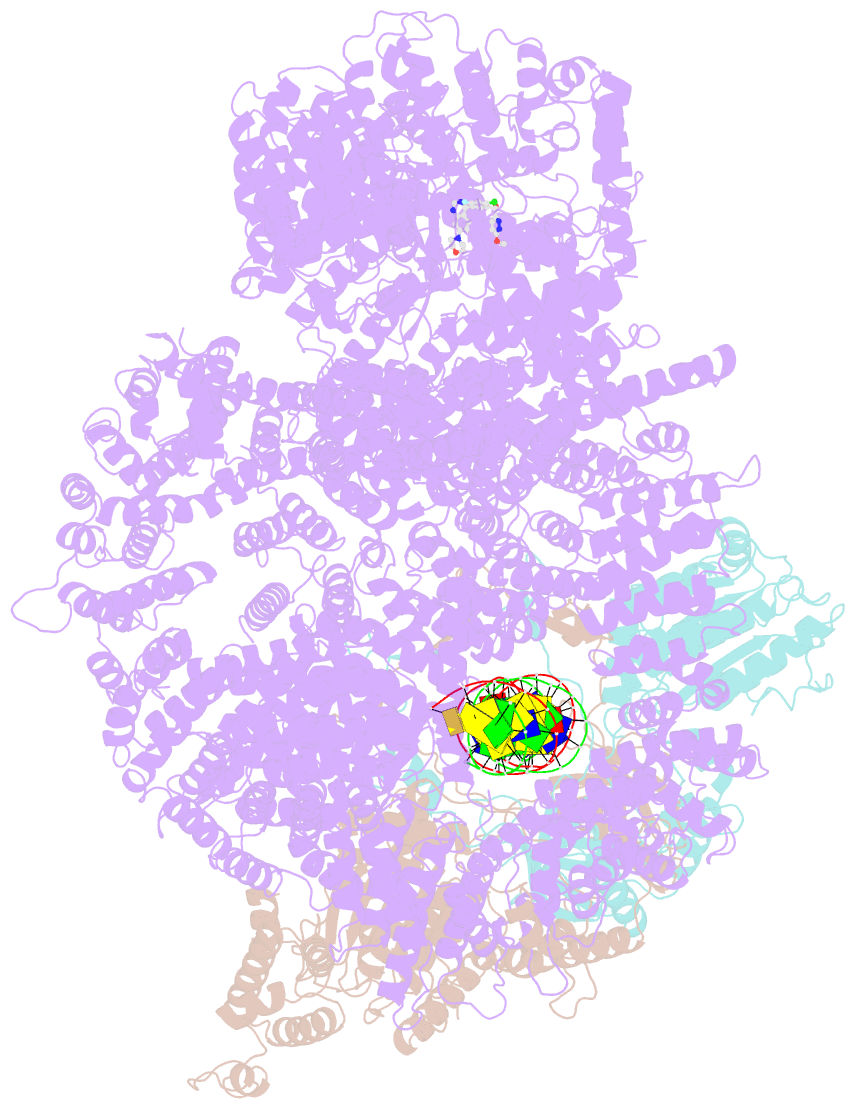
- DNA-pk in the active state
- Reference
- Liang S, Blundell TL (2023): "Human DNA-dependent protein kinase activation mechanism." Nat.Struct.Mol.Biol., 30, 140-147. doi: 10.1038/s41594-022-00881-w.
- Abstract
- DNA-dependent protein kinase (DNA-PK), a multicomponent complex including the DNA-PK catalytic subunit and Ku70/80 heterodimer together with DNA, is central to human DNA damage response and repair. Using a DNA-PK-selective inhibitor (M3814), we identified from one dataset two cryo-EM structures of the human DNA-PK complex in different states, the intermediate state and the active state. Here we show that activation of the kinase is regulated through conformational changes caused by the binding ligand and the string region (residues 802-846) of the DNA-PK catalytic subunit, particularly the helix-hairpin-helix motif (residues 816-836) that interacts with DNA. These observations demonstrate the regulatory role of the ligand and explain why DNA-PK is DNA dependent. Cooperation and coordination among binding partners, disordered flexible regions and mechanically flexible HEAT repeats modulate the activation of the kinase. Together with previous findings, these results provide a better molecular understanding of DNA-PK catalysis.