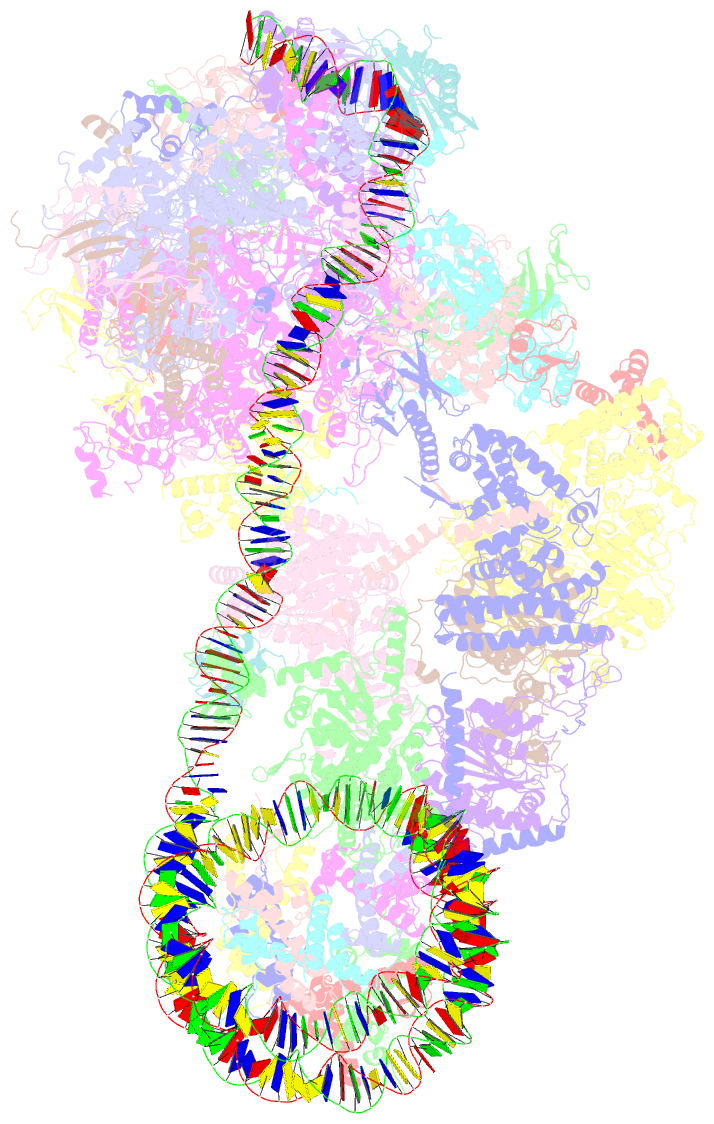
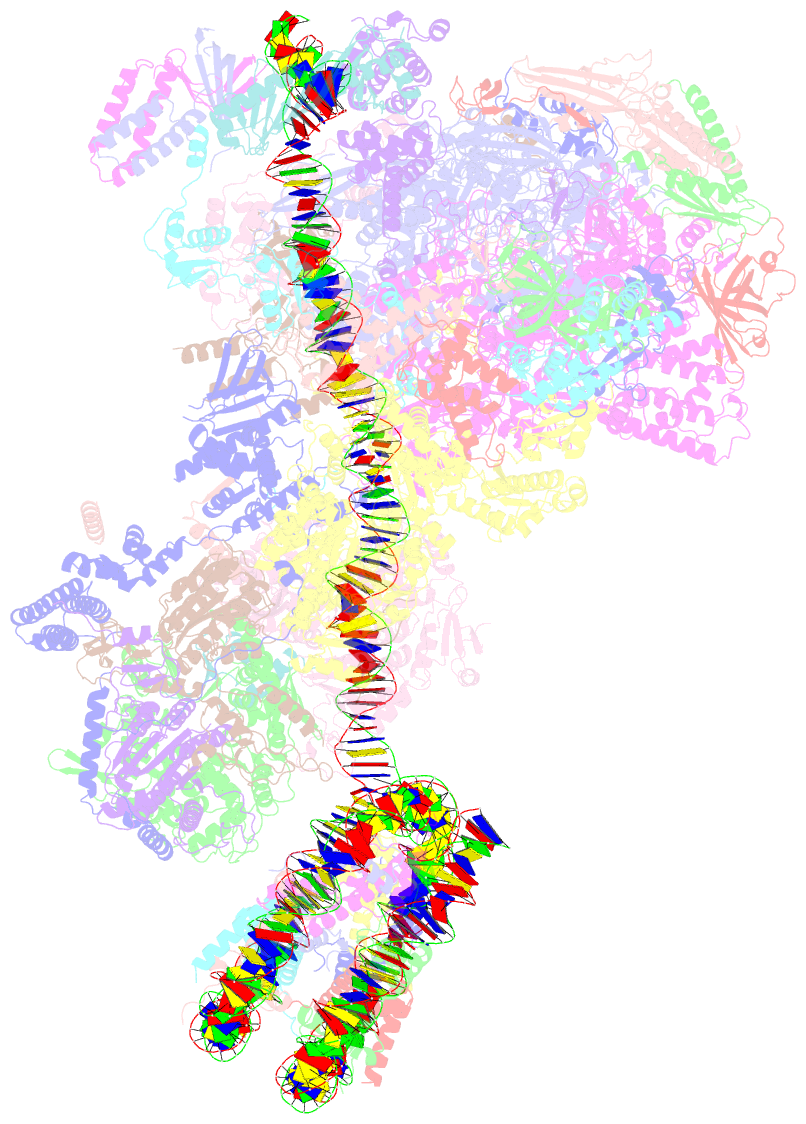
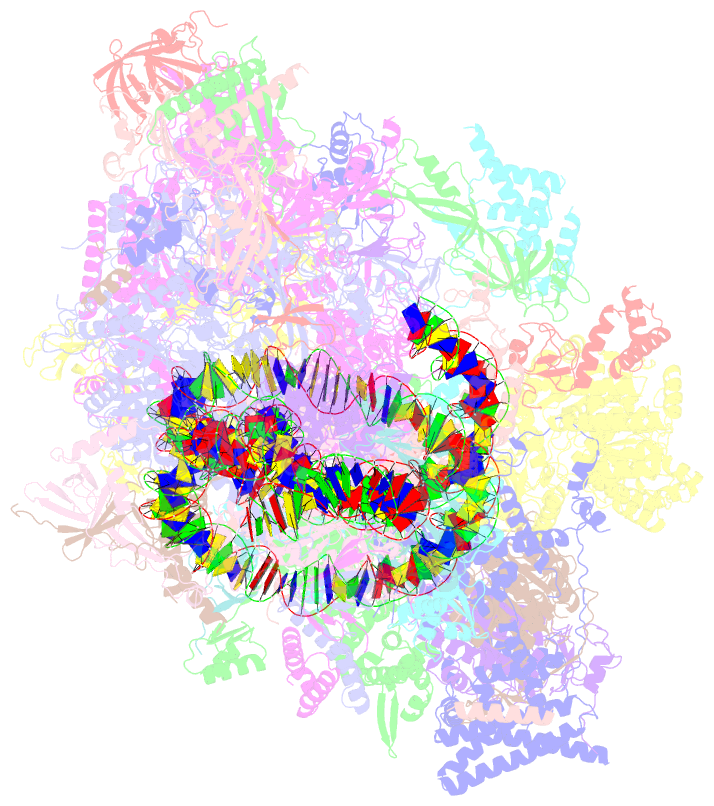
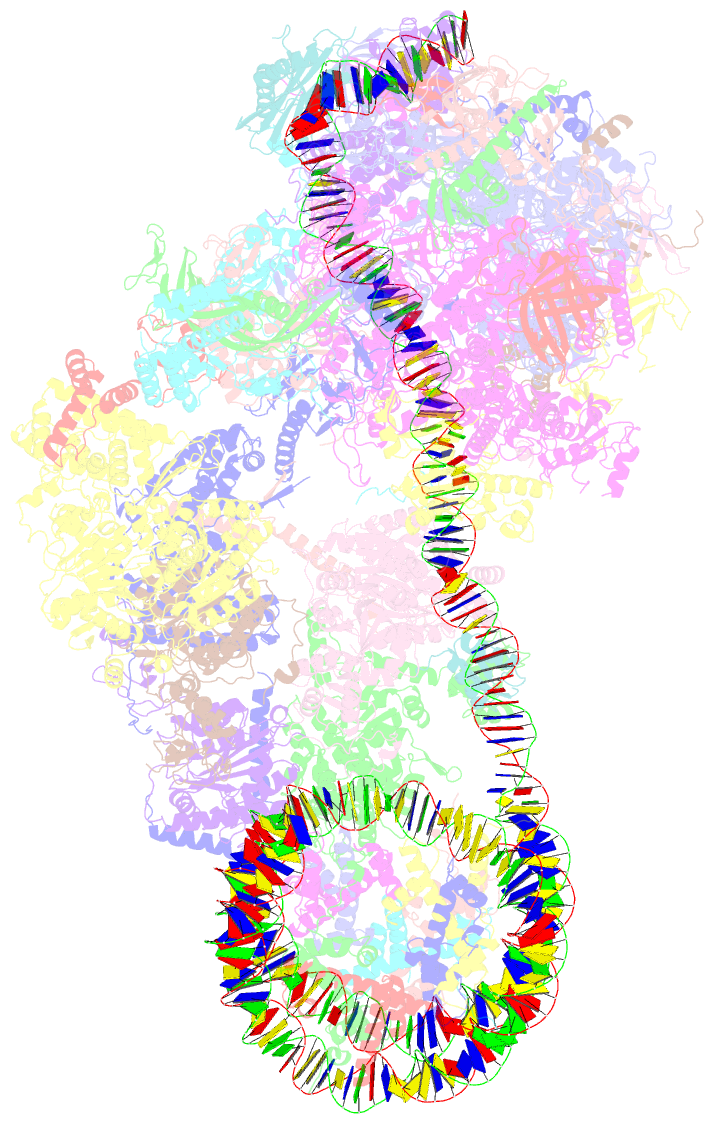
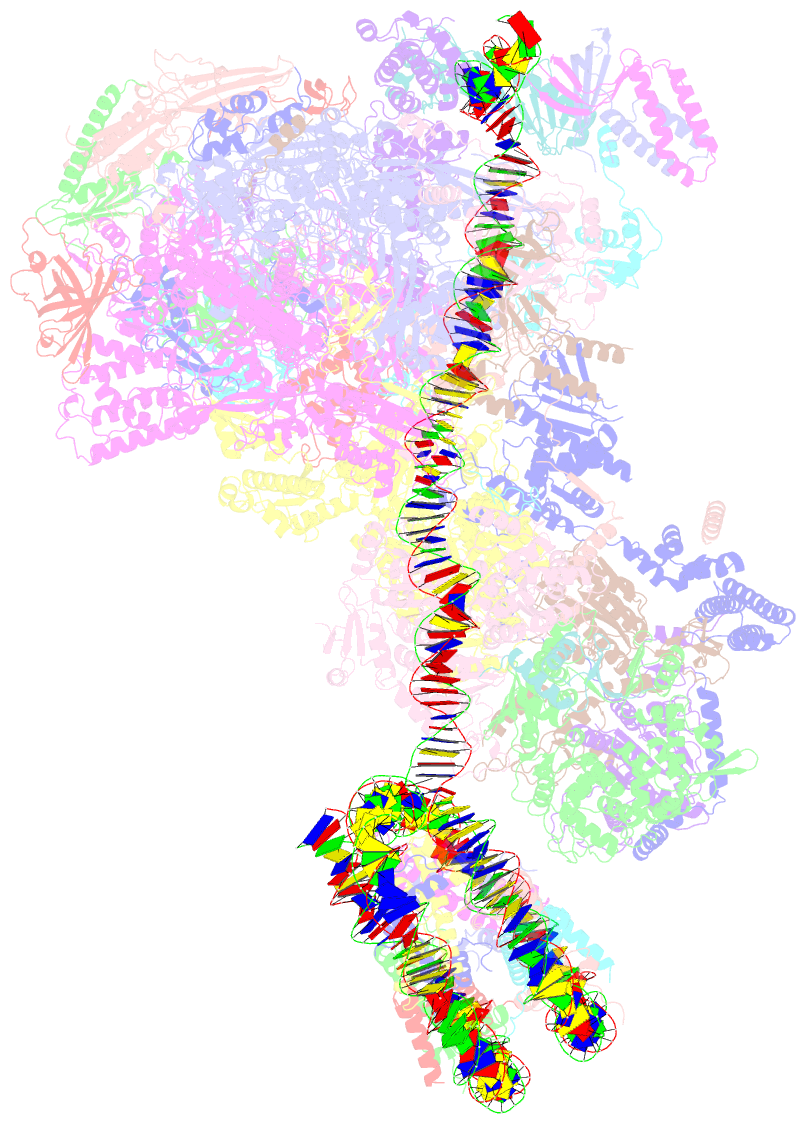
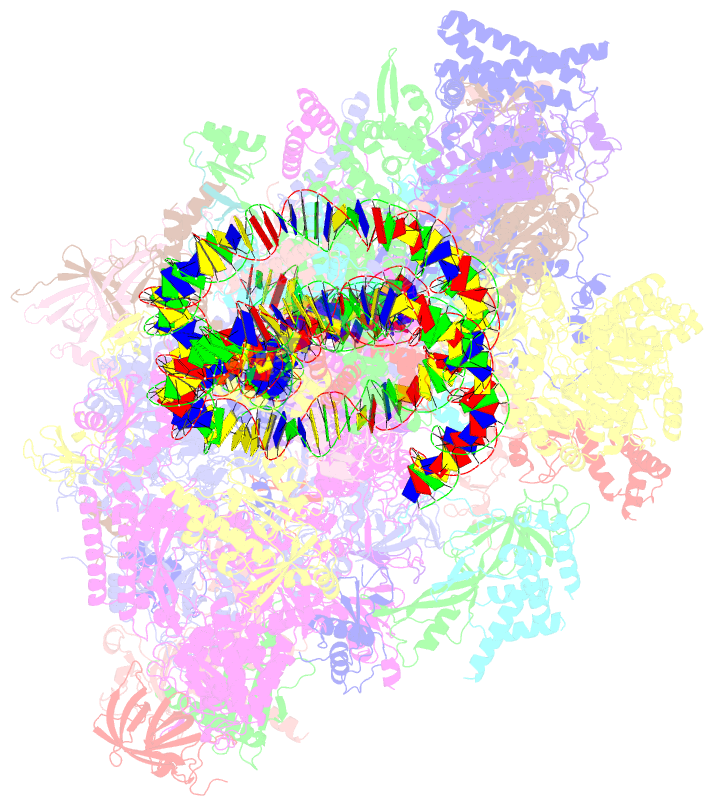
Summary information and primary citation
- PDB-id
- 7zsb; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription
- Method
- cryo-EM (6.6 Å)
- Summary
- Yeast RNA polymerase ii transcription pre-initiation complex with the +1 nucleosome and ntp, complex c
- Reference
- Wang H, Schilbach S, Ninov M, Urlaub H, Cramer P (2023): "Structures of transcription preinitiation complex engaged with the +1 nucleosome." Nat.Struct.Mol.Biol., 30, 226-232. doi: 10.1038/s41594-022-00865-w.
- Abstract
- The preinitiation complex (PIC) assembles on promoters of protein-coding genes to position RNA polymerase II (Pol II) for transcription initiation. Previous structural studies revealed the PIC on different promoters, but did not address how the PIC assembles within chromatin. In the yeast Saccharomyces cerevisiae, PIC assembly occurs adjacent to the +1 nucleosome that is located downstream of the core promoter. Here we present cryo-EM structures of the yeast PIC bound to promoter DNA and the +1 nucleosome located at three different positions. The general transcription factor TFIIH engages with the incoming downstream nucleosome and its translocase subunit Ssl2 (XPB in human TFIIH) drives the rotation of the +1 nucleosome leading to partial detachment of nucleosomal DNA and intimate interactions between TFIIH and the nucleosome. The structures provide insights into how transcription initiation can be influenced by the +1 nucleosome and may explain why the transcription start site is often located roughly 60 base pairs upstream of the dyad of the +1 nucleosome in yeast.