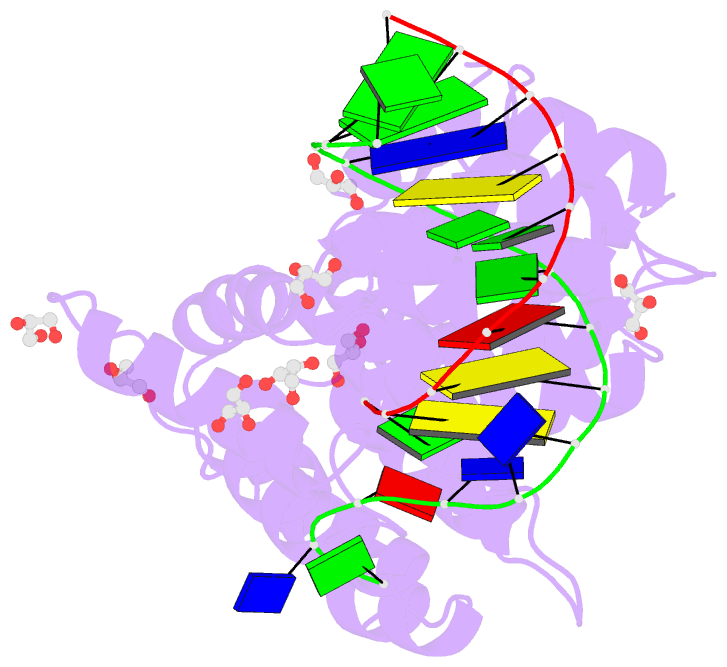
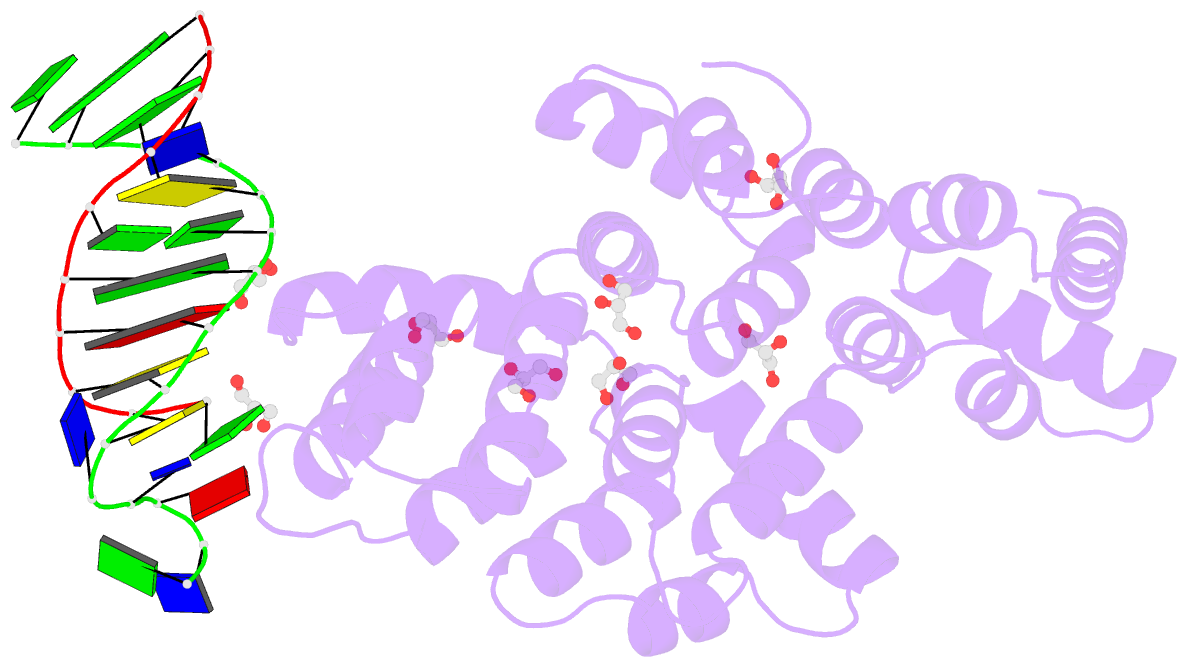
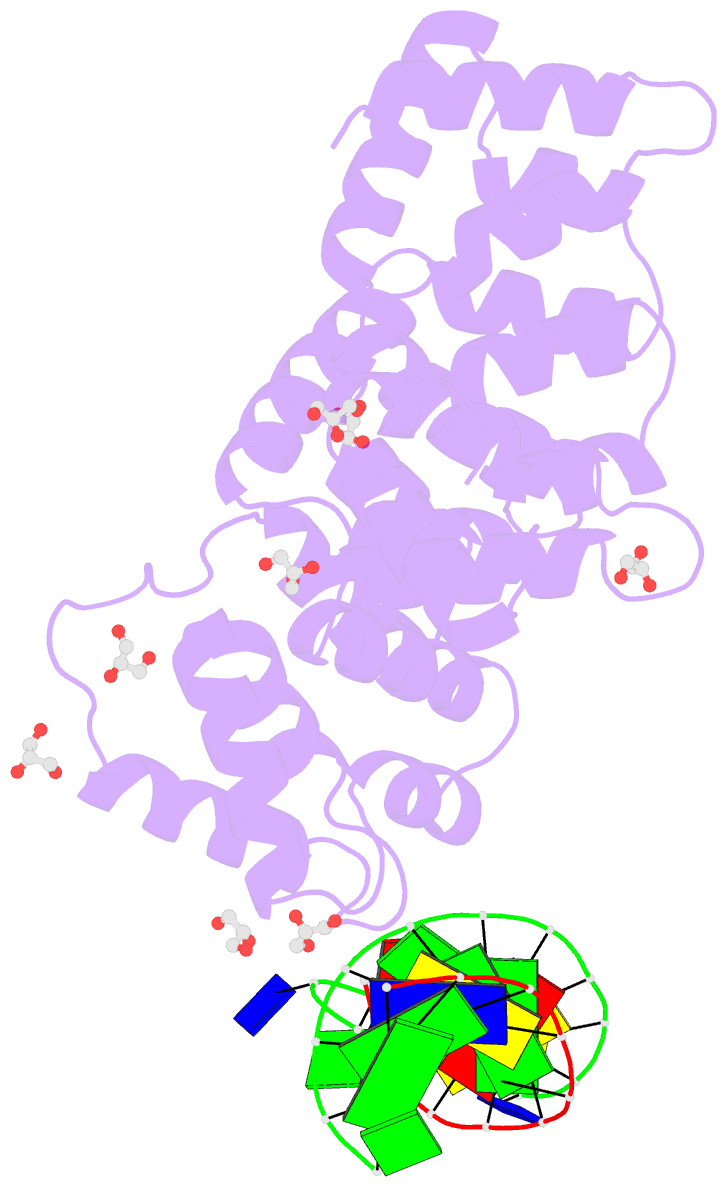
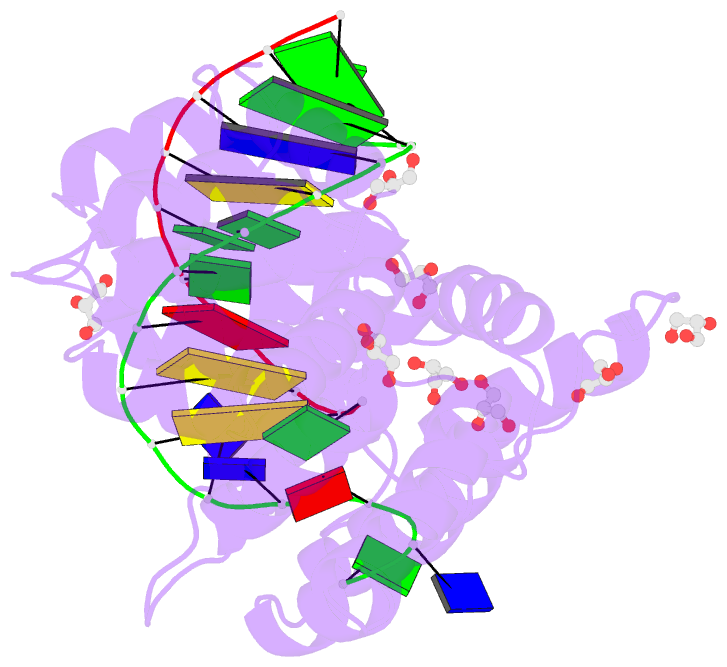
Summary information and primary citation
- PDB-id
- 7zvn; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- lipid binding protein
- Method
- X-ray (1.87 Å)
- Summary
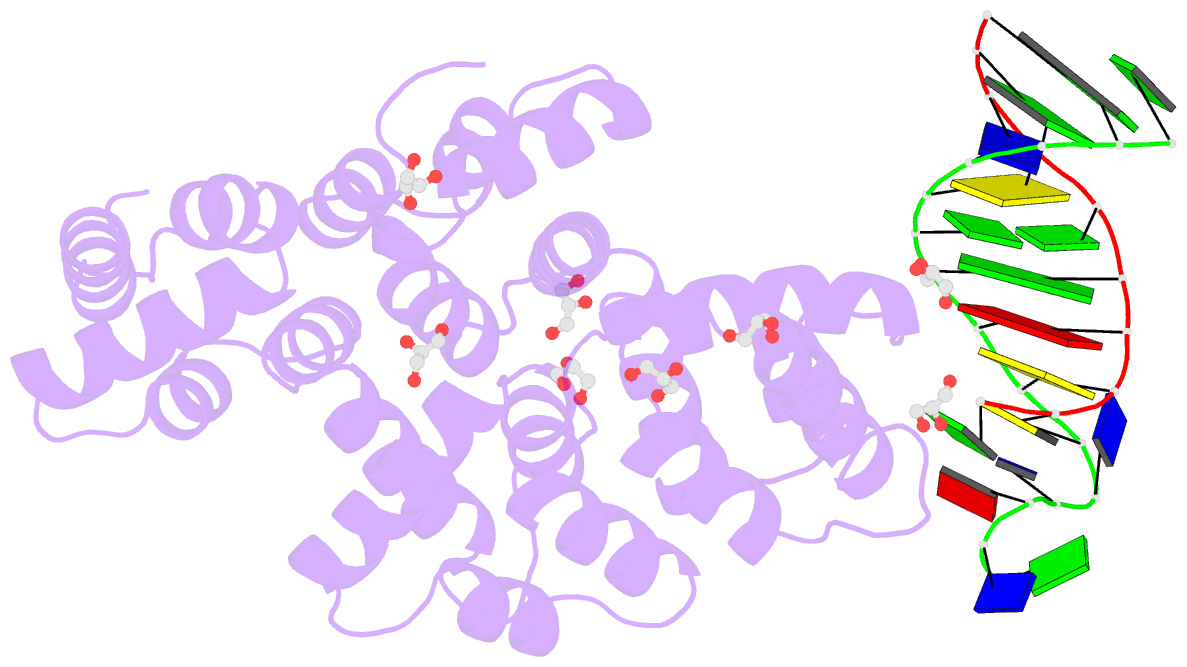
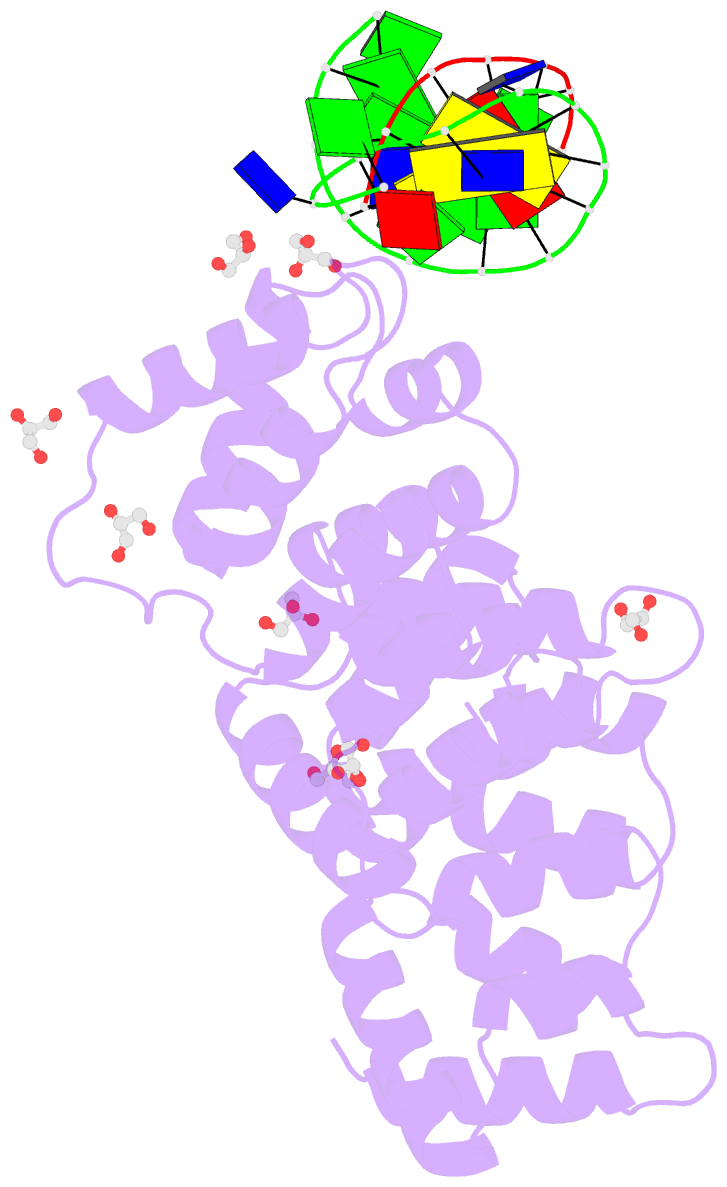
- Crystal structure of human annexin a2 in complex with full phosphorothioate 5-10 2'-methoxyethyl DNA gapmer antisense oligonucleotide solved at 1.87 Å resolution
- Reference
- Hyjek-Skladanowska M, Anderson BA, Mykhaylyk V, Orr C, Wagner A, Poznanski JT, Skowronek K, Seth P, Nowotny M (2023): "Structures of annexin A2-PS DNA complexes show dominance of hydrophobic interactions in phosphorothioate binding." Nucleic Acids Res., 51, 1409-1423. doi: 10.1093/nar/gkac774.
- Abstract
- The introduction of phosphorothioate (PS) linkages to the backbone of therapeutic nucleic acids substantially increases their stability and potency. It also affects their interactions with cellular proteins, but the molecular mechanisms that underlie this effect are poorly understood. Here, we report structural and biochemical studies of interactions between annexin A2, a protein that does not possess any known canonical DNA binding domains, and phosphorothioate-modified antisense oligonucleotides. We show that a unique mode of hydrophobic interactions between a sulfur atom of the phosphorothioate group and lysine and arginine residues account for the enhanced affinity of modified nucleic acid for the protein. Our results demonstrate that this mechanism of interaction is observed not only for nucleic acid-binding proteins but can also account for the association of PS oligonucleotides with other proteins. Using the anomalous diffraction of sulfur, we showed that preference for phosphorothioate stereoisomers is determined by the hydrophobic environment around the PS linkage that comes not only from protein but also from additional structural features within the ASO such as 5-Me groups on cytosine nucleobases.