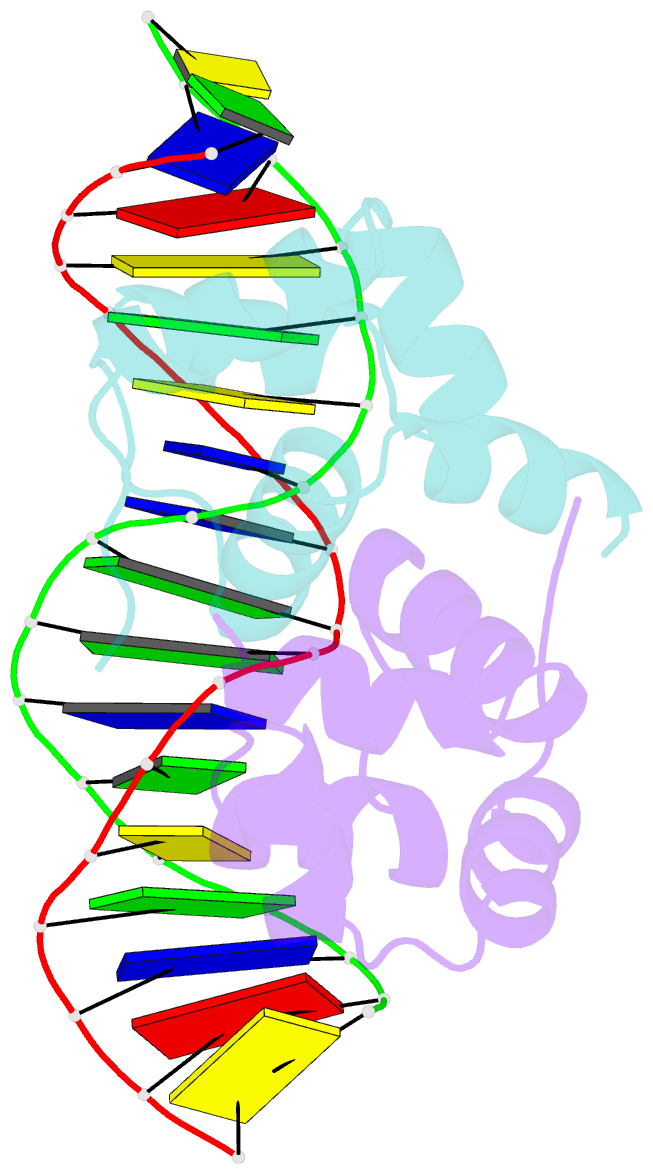
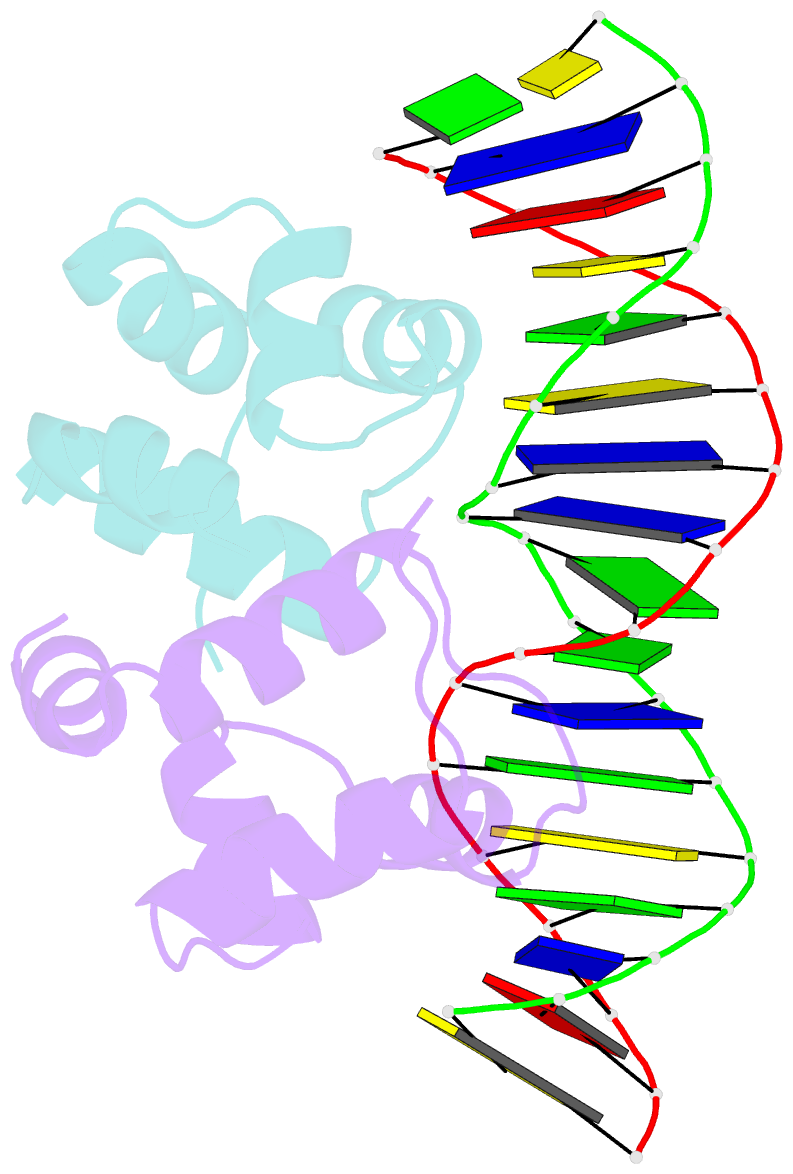
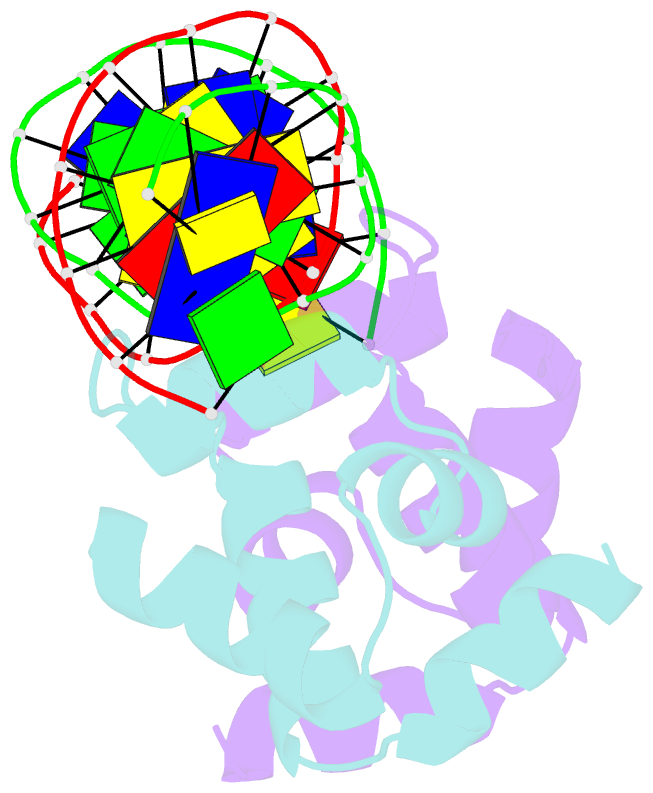
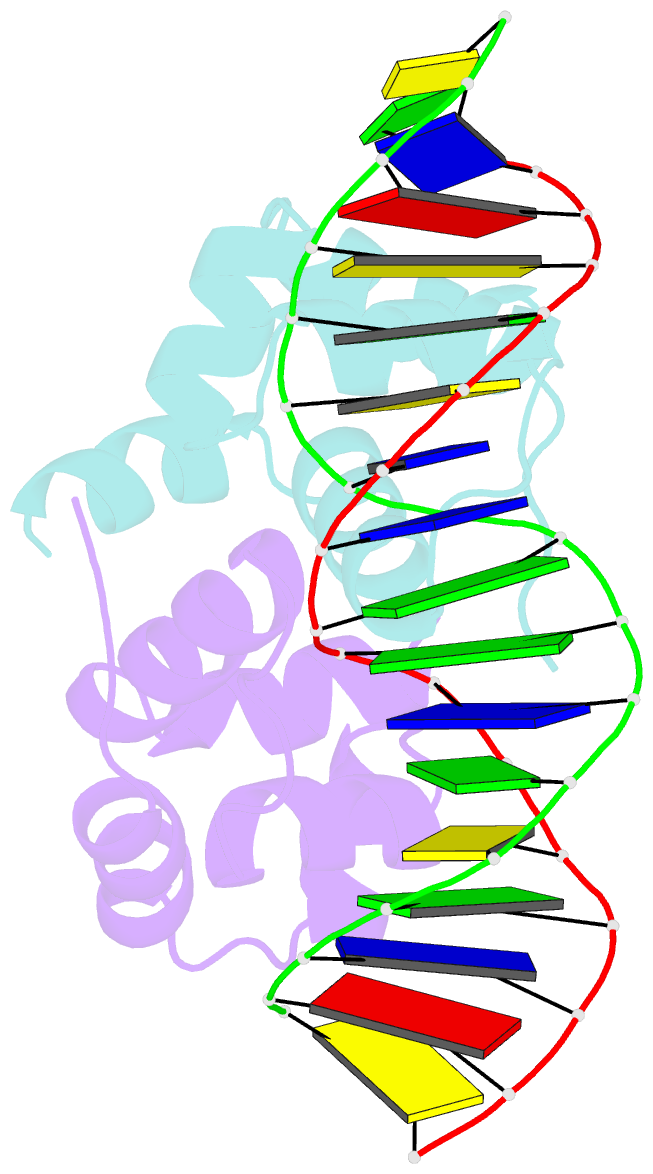
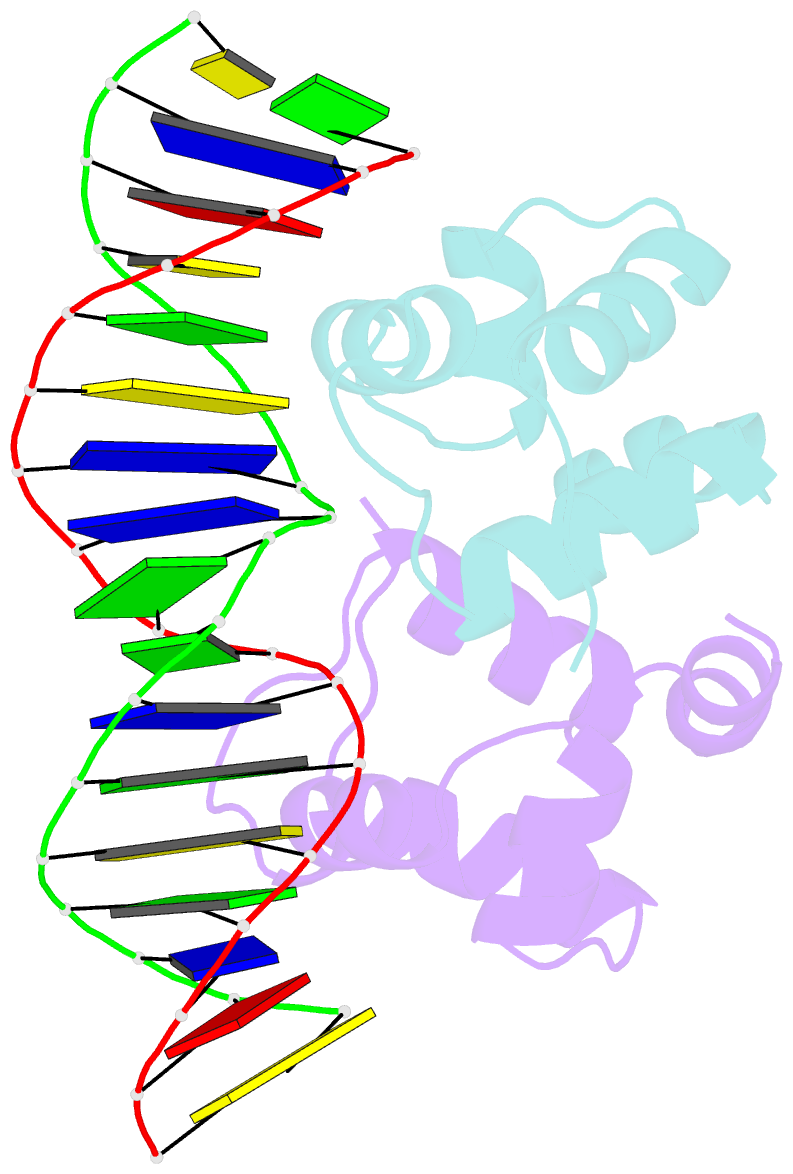
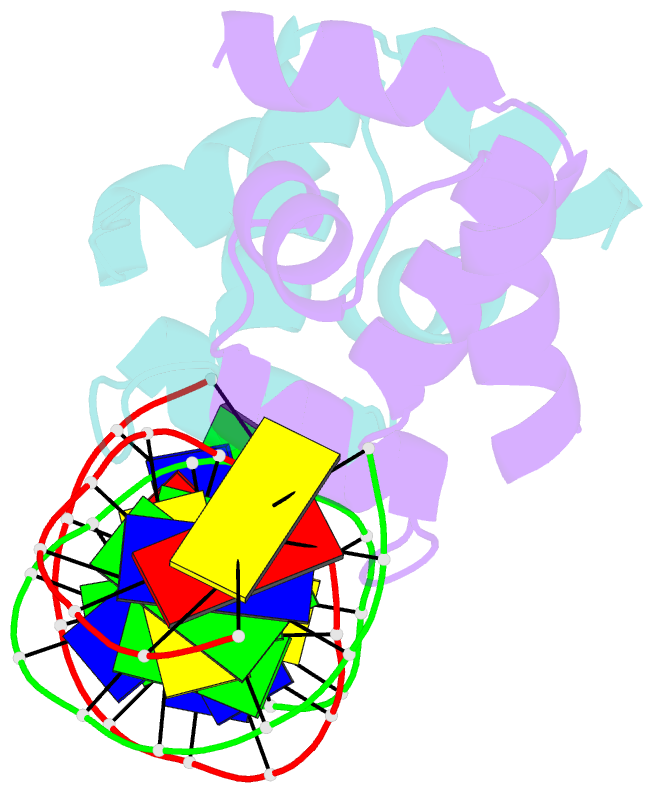
Summary information and primary citation
- PDB-id
- 8a0w; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- DNA binding protein
- Method
- X-ray (2.334 Å)
- Summary
- Crystal structure of the higa2 antitoxin in complex with operator DNA
- Reference
- Hadzi S, Zivic Z, Kovacic M, Zavrtanik U, Haeserts S, Charlier D, Plavec J, Volkov AN, Lah J, Loris R (2024): "Fuzzy recognition by the prokaryotic transcription factor HigA2 from Vibrio cholerae." Nat Commun, 15, 3105. doi: 10.1038/s41467-024-47296-3.
- Abstract
- Disordered protein sequences can exhibit different binding modes, ranging from well-ordered folding-upon-binding to highly dynamic fuzzy binding. The primary function of the intrinsically disordered region of the antitoxin HigA2 from Vibrio cholerae is to neutralize HigB2 toxin through ultra-high-affinity folding-upon-binding interaction. Here, we show that the same intrinsically disordered region can also mediate fuzzy interactions with its operator DNA and, through interplay with the folded helix-turn-helix domain, regulates transcription from the higBA2 operon. NMR, SAXS, ITC and in vivo experiments converge towards a consistent picture where a specific set of residues in the intrinsically disordered region mediate electrostatic and hydrophobic interactions while "hovering" over the DNA operator. Sensitivity of the intrinsically disordered region to scrambling the sequence, position-specific contacts and absence of redundant, multivalent interactions, point towards a more specific type of fuzzy binding. Our work demonstrates how a bacterial regulator achieves dual functionality by utilizing two distinct interaction modes within the same disordered sequence.