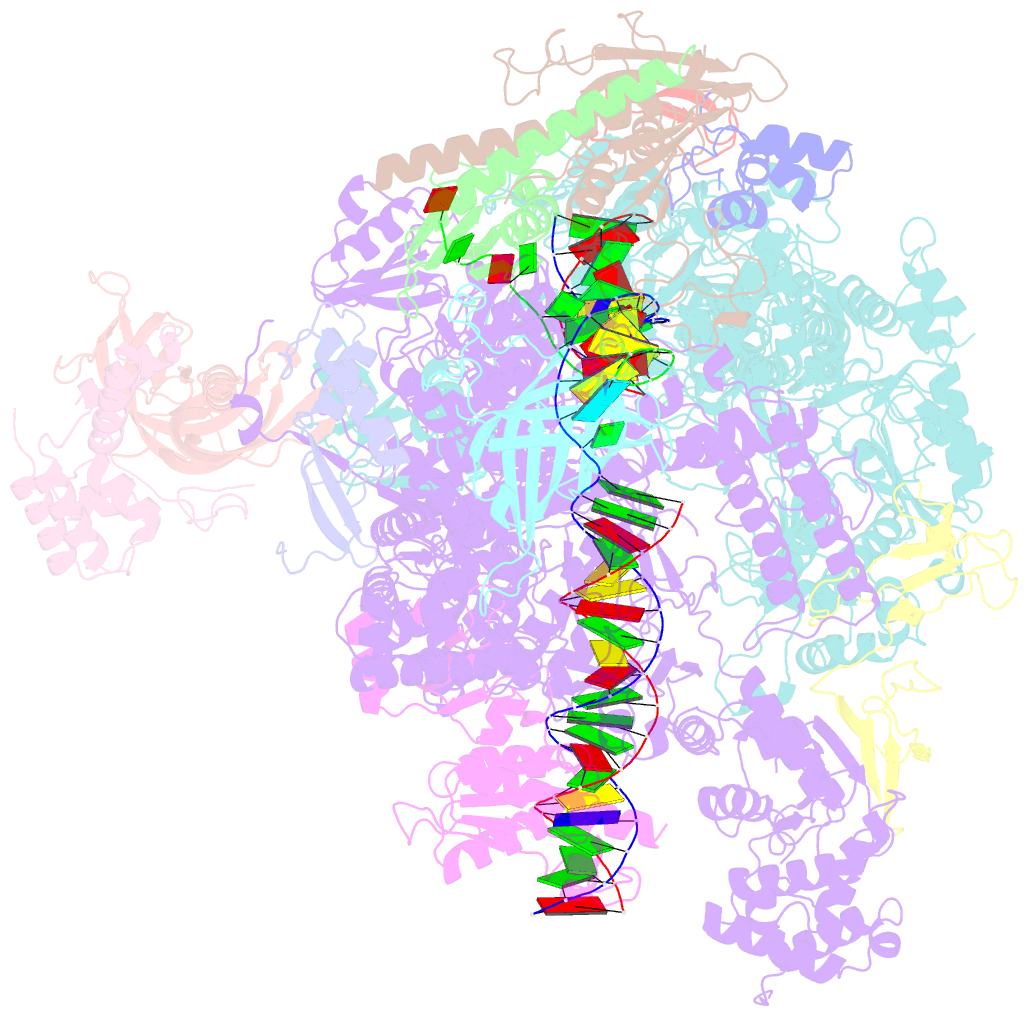
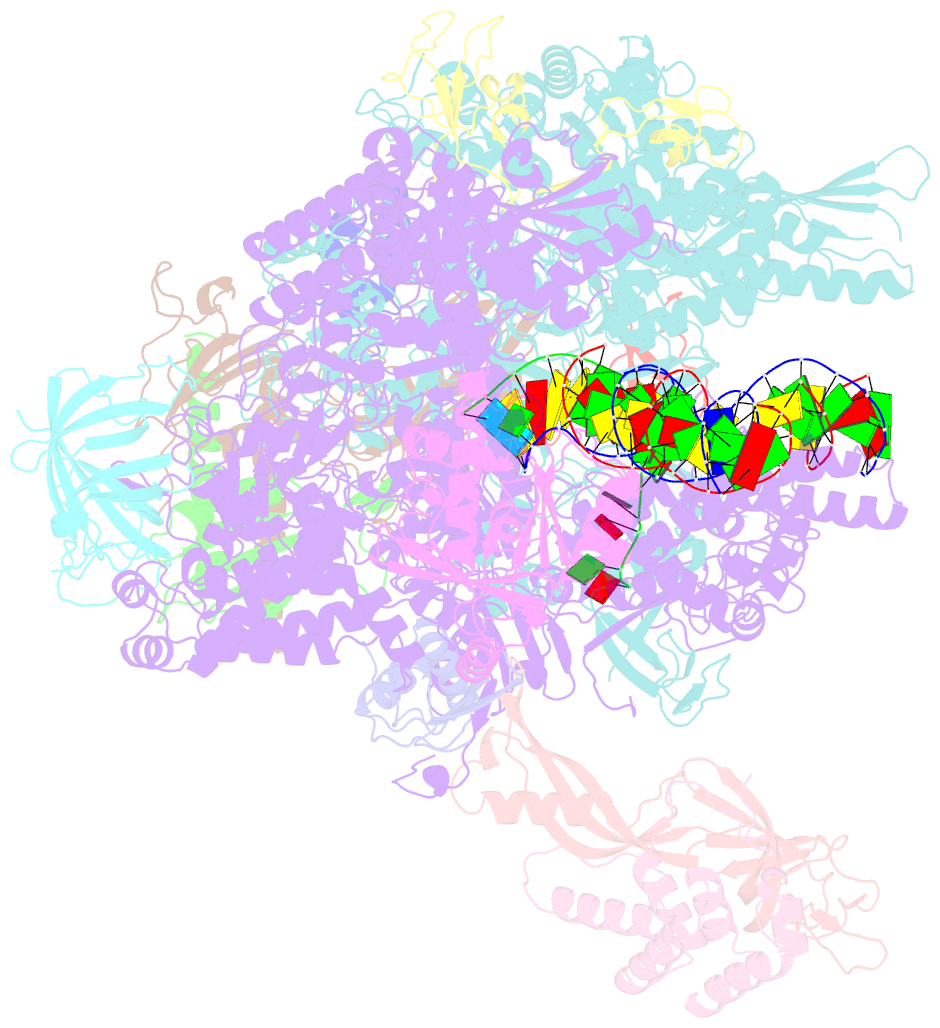
Summary information and primary citation
- PDB-id
- 8a40; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription
- Method
- cryo-EM (3.0 Å)
- Summary
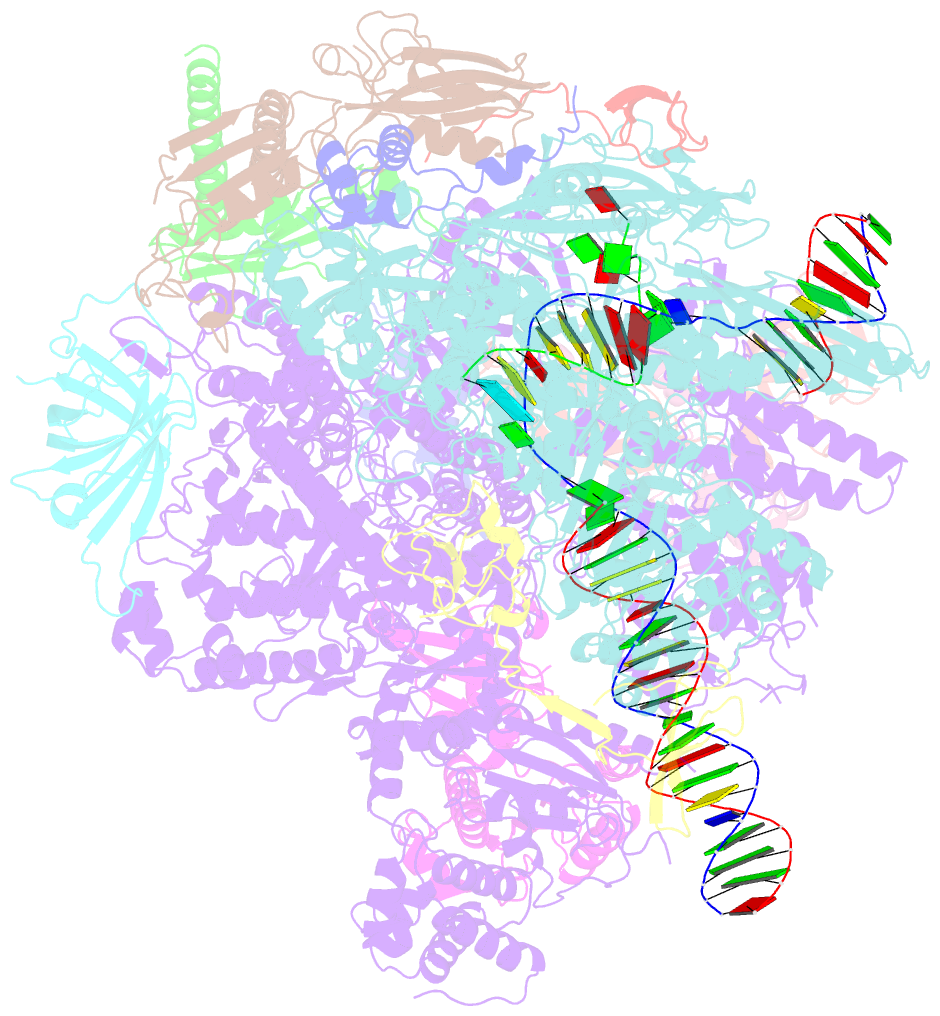
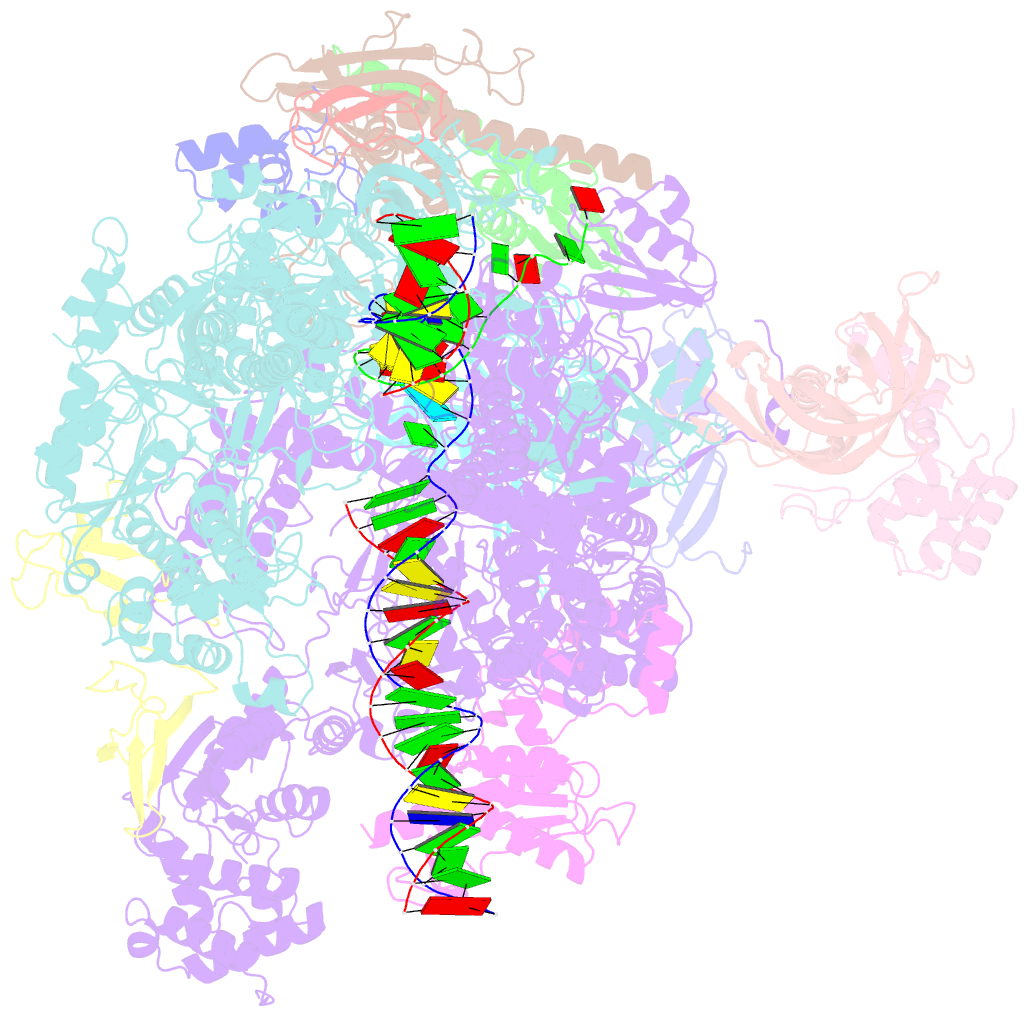
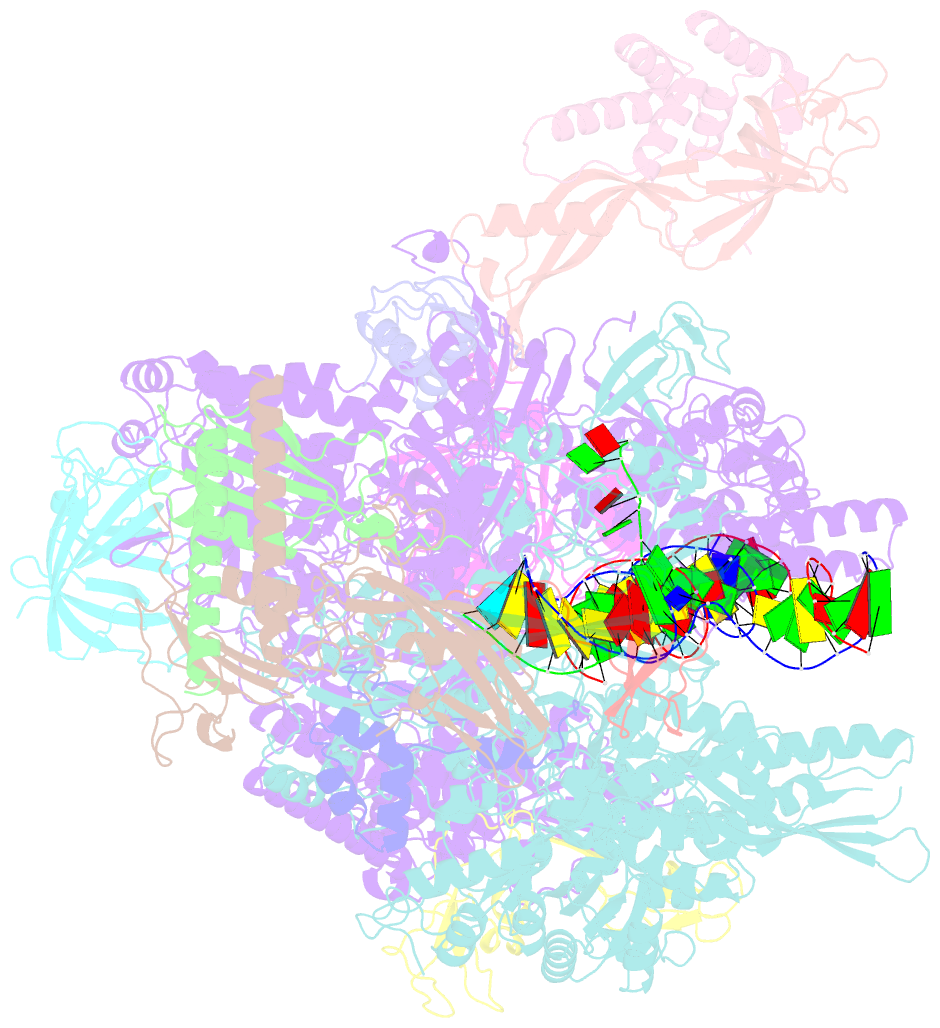
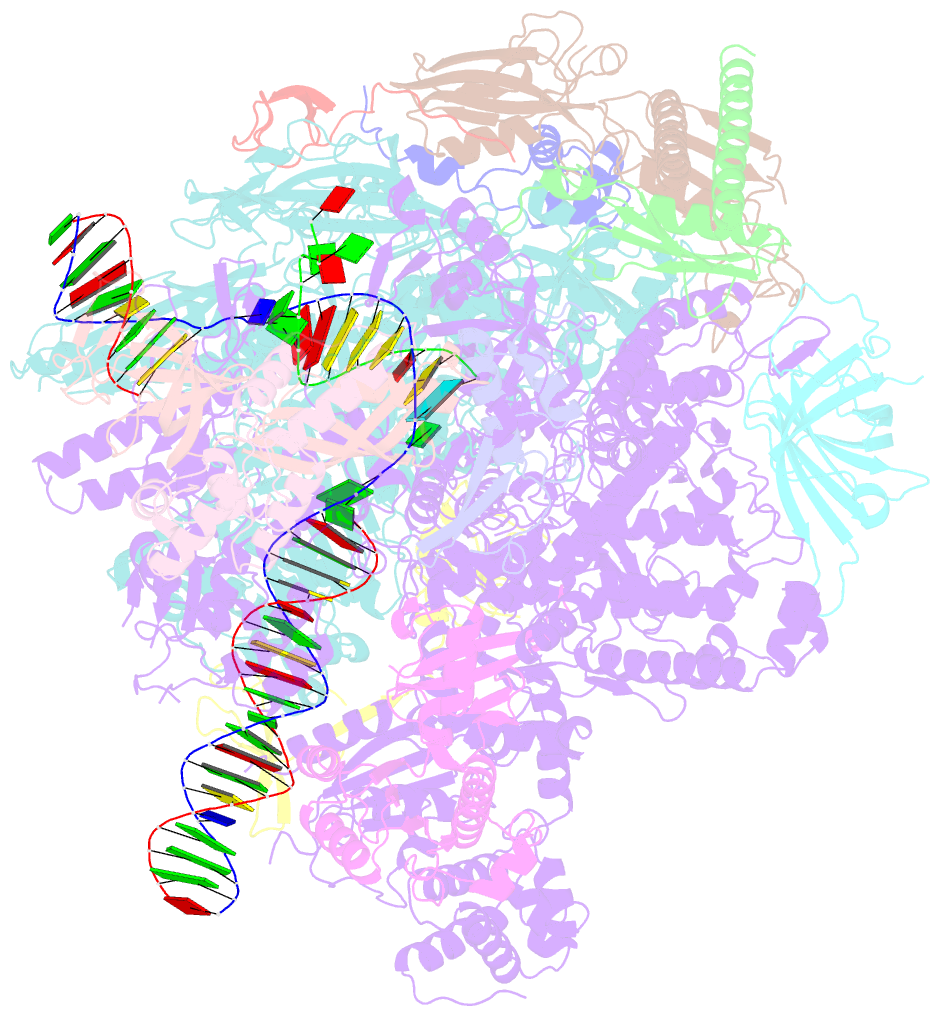
- Structure of mammalian pol ii-tfiis elongation complex
- Reference
- Farnung L, Ochmann M, Garg G, Vos SM, Cramer P (2022): "Structure of a backtracked hexasomal intermediate of nucleosome transcription." Mol.Cell, 82, 3126-3134.e7. doi: 10.1016/j.molcel.2022.06.027.
- Abstract
- During gene transcription, RNA polymerase II (RNA Pol II) passes nucleosomes with the help of various elongation factors. Here, we show that RNA Pol II achieves efficient nucleosome passage when the human elongation factors DSIF, PAF1 complex (PAF), RTF1, SPT6, and TFIIS are present. The cryo-EM structure of an intermediate of the nucleosome passage shows a partially unraveled hexasome that lacks the proximal H2A-H2B dimer and interacts with the RNA Pol II jaw, DSIF, and the CTR9trestle helix. RNA Pol II adopts a backtracked state with the RNA 3' end dislodged from the active site and bound in the RNA Pol II pore. Additional structures and biochemical data show that human TFIIS enters the RNA Pol II pore and stimulates the cleavage of the backtracked RNA and nucleosome passage.