Summary information and primary citation
- PDB-id
- 8a4i; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- DNA binding protein
- Method
- X-ray (2.76 Å)
- Summary
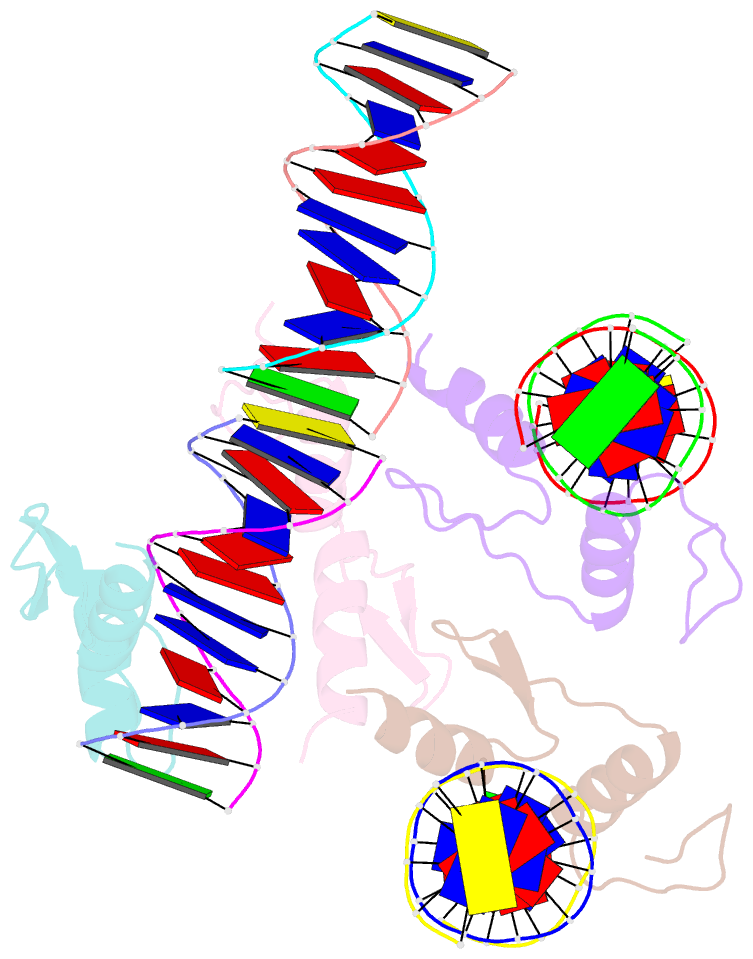
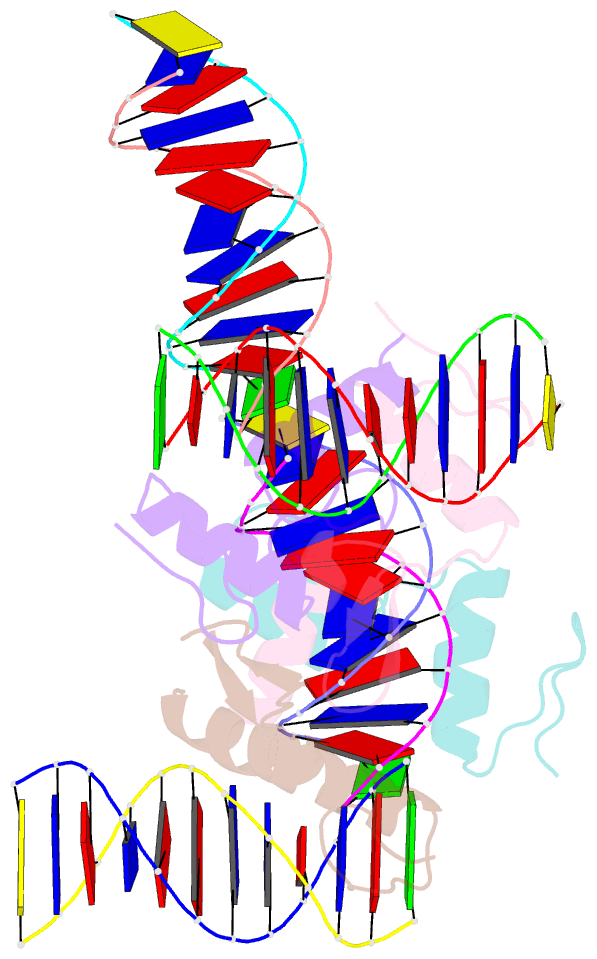
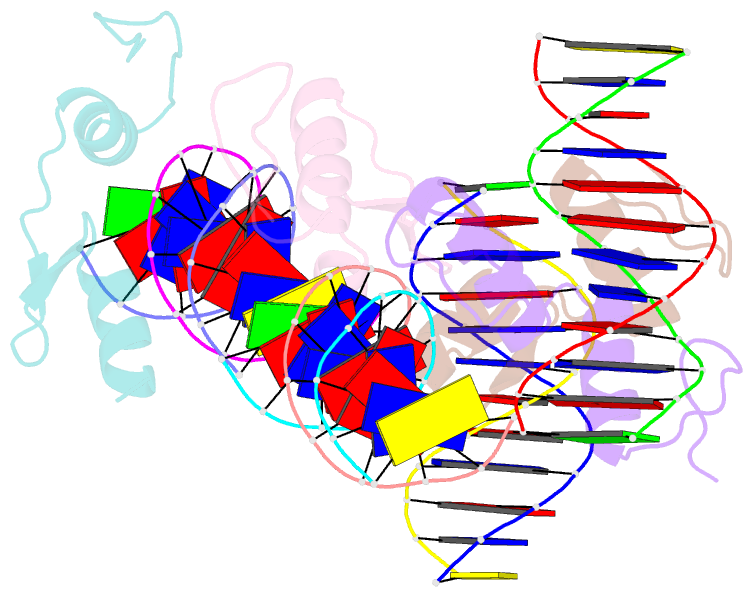
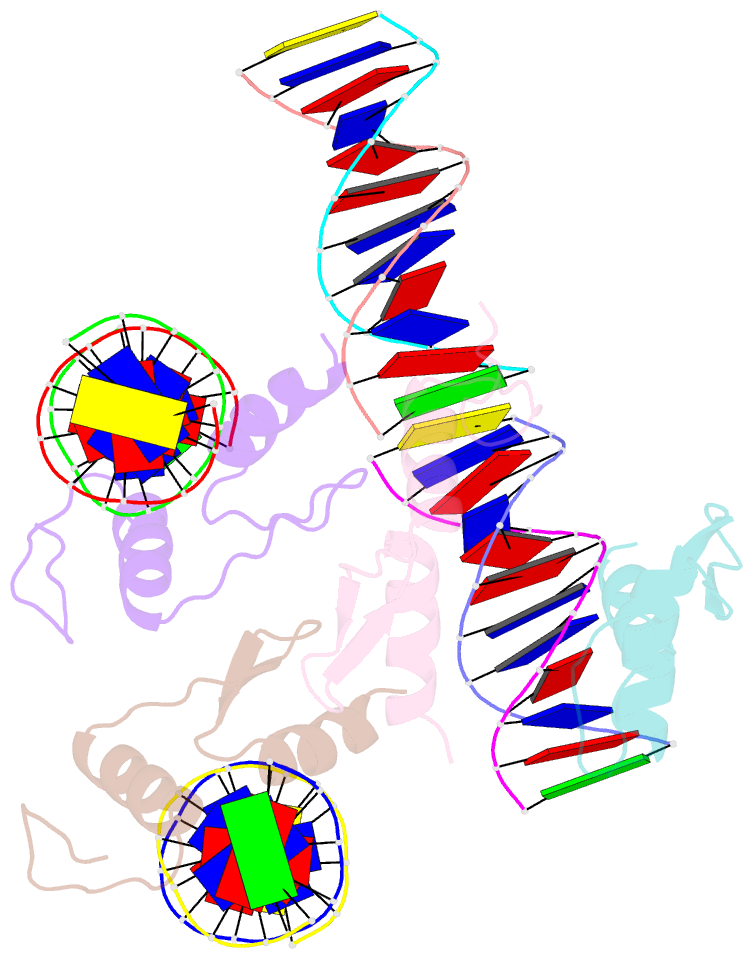
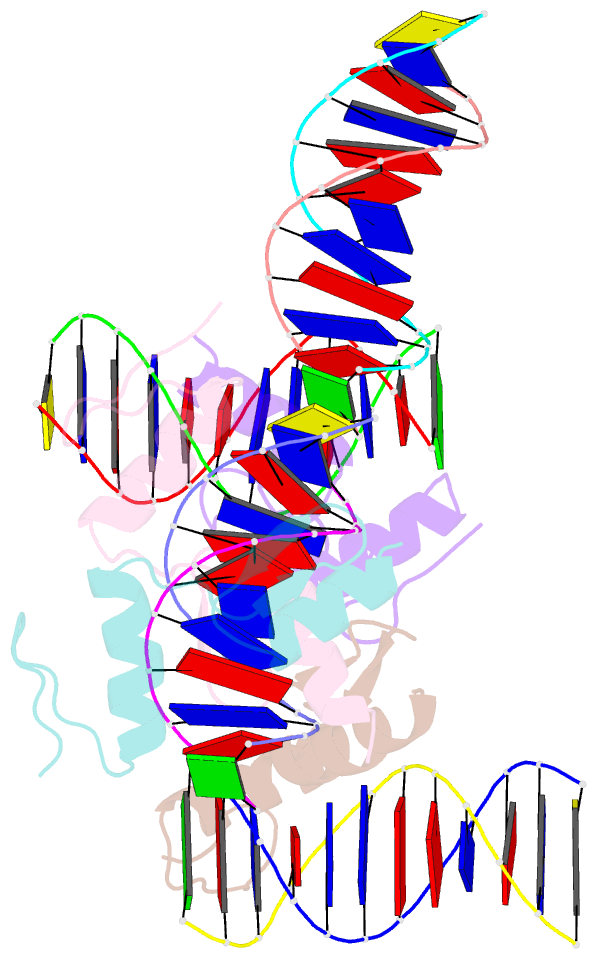
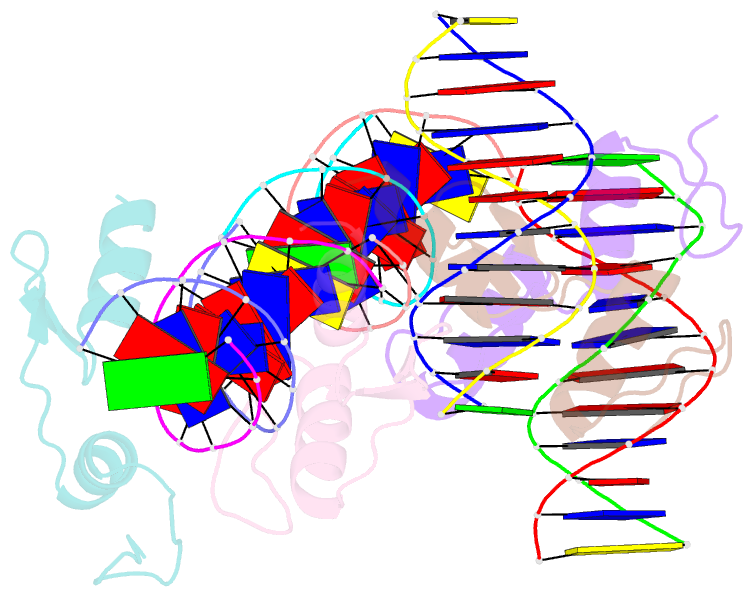
- Crystal structure of sall4 zinc finger cluster 4 with at-rich DNA
- Reference
- Watson JA, Pantier R, Jayachandran U, Chhatbar K, Alexander-Howden B, Kruusvee V, Prendecki M, Bird A, Cook AG (2023): "Structure of SALL4 zinc finger domain reveals link between AT-rich DNA binding and Okihiro syndrome." Life Sci Alliance, 6. doi: 10.26508/lsa.202201588.
- Abstract
- Spalt-like 4 (SALL4) maintains vertebrate embryonic stem cell identity and is required for the development of multiple organs, including limbs. Mutations in SALL4 are associated with Okihiro syndrome, and SALL4 is also a known target of thalidomide. SALL4 protein has a distinct preference for AT-rich sequences, recognised by a pair of zinc fingers at the C-terminus. However, unlike many characterised zinc finger proteins, SALL4 shows flexible recognition with many different combinations of AT-rich sequences being targeted. SALL4 interacts with the NuRD corepressor complex which potentially mediates repression of AT-rich genes. We present a crystal structure of SALL4 C-terminal zinc fingers with an AT-rich DNA sequence, which shows that SALL4 uses small hydrophobic and polar side chains to provide flexible recognition in the major groove. Missense mutations reported in patients that lie within the C-terminal zinc fingers reduced overall binding to DNA but not the preference for AT-rich sequences. Furthermore, these mutations altered association of SALL4 with AT-rich genomic sites, providing evidence that these mutations are likely pathogenic.