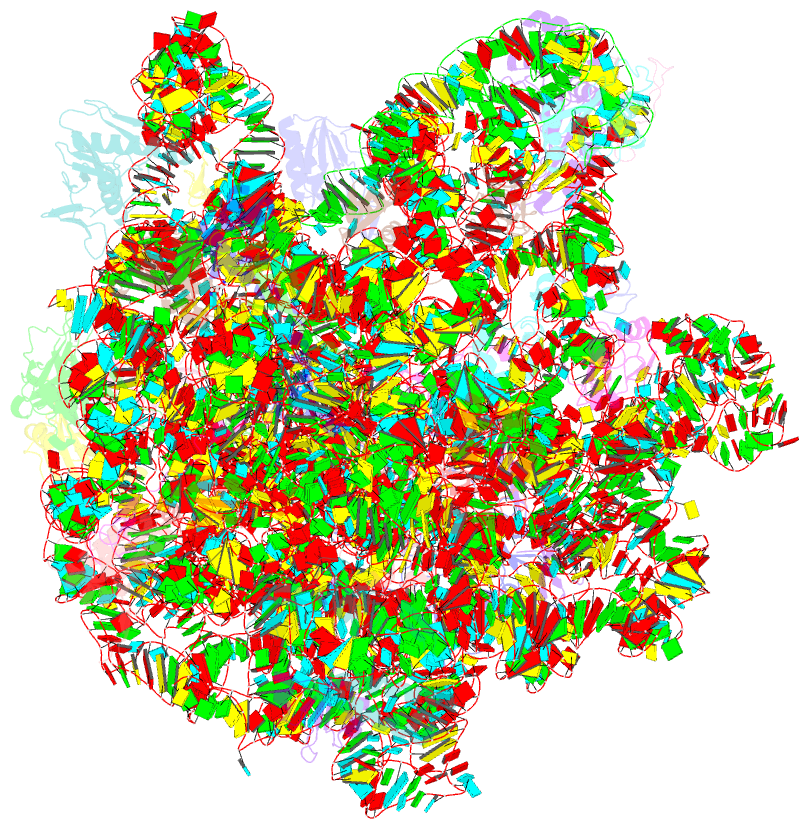
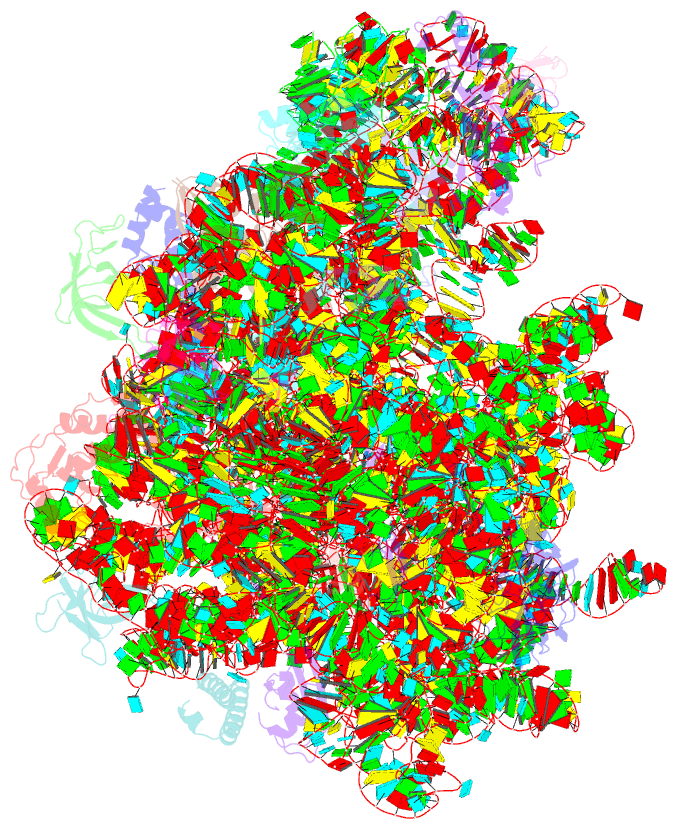
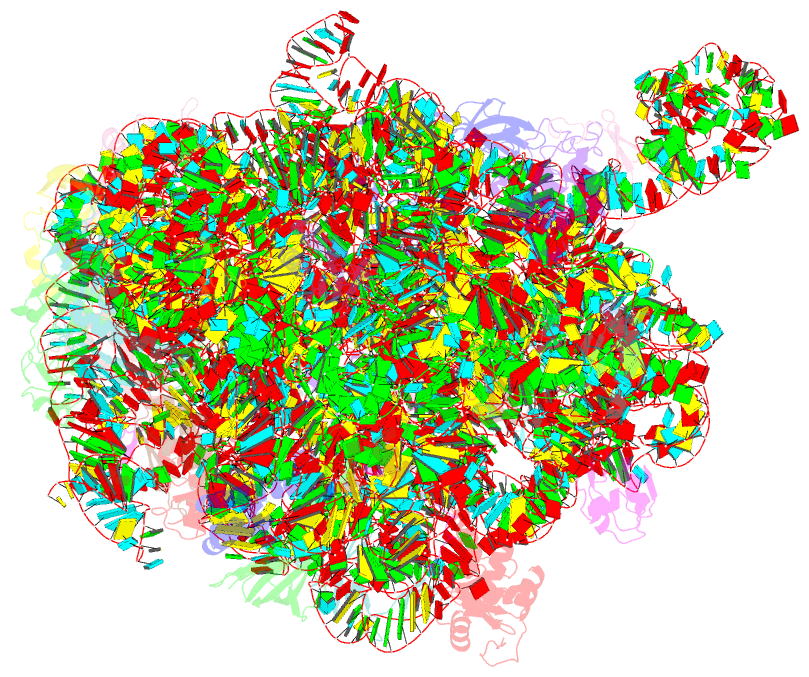
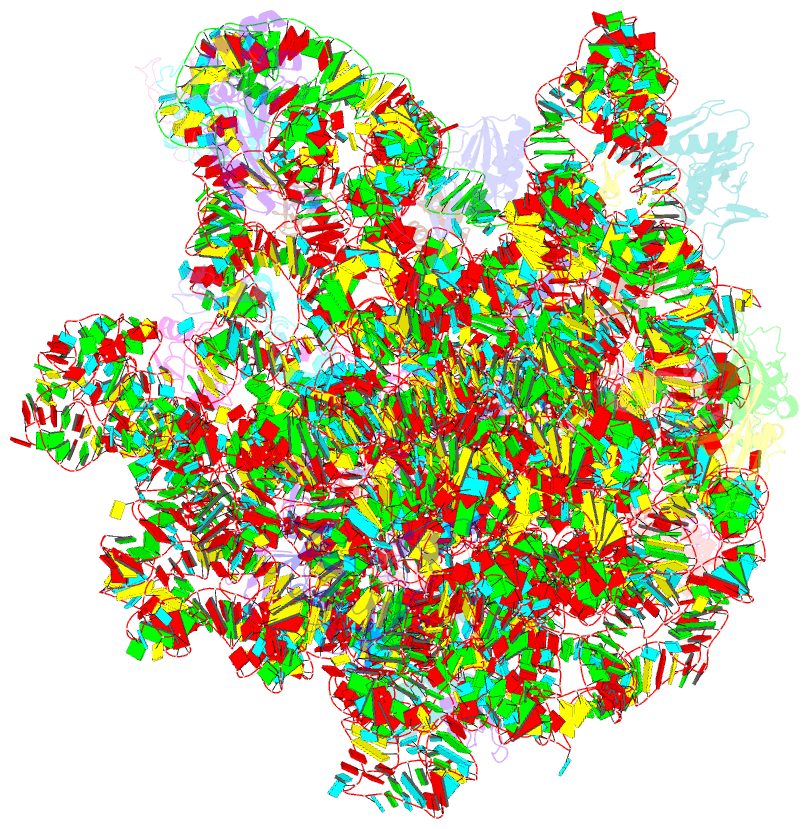
Summary information and primary citation
- PDB-id
- 8a5i; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- ribosome
- Method
- cryo-EM (2.3 Å)
- Summary
- cryo-EM structure of lincomycin bound to the listeria monocytogenes 50s ribosomal subunit.
- Reference
- Koller TO, Turnbull KJ, Vaitkevicius K, Crowe-McAuliffe C, Roghanian M, Bulvas O, Nakamoto JA, Kurata T, Julius C, Atkinson GC, Johansson J, Hauryliuk V, Wilson DN (2022): "Structural basis for HflXr-mediated antibiotic resistance in Listeria monocytogenes." Nucleic Acids Res., 50, 11285-11300. doi: 10.1093/nar/gkac934.
- Abstract
- HflX is a ubiquitous bacterial GTPase that splits and recycles stressed ribosomes. In addition to HflX, Listeria monocytogenes contains a second HflX homolog, HflXr. Unlike HflX, HflXr confers resistance to macrolide and lincosamide antibiotics by an experimentally unexplored mechanism. Here, we have determined cryo-EM structures of L. monocytogenes HflXr-50S and HflX-50S complexes as well as L. monocytogenes 70S ribosomes in the presence and absence of the lincosamide lincomycin. While the overall geometry of HflXr on the 50S subunit is similar to that of HflX, a loop within the N-terminal domain of HflXr, which is two amino acids longer than in HflX, reaches deeper into the peptidyltransferase center. Moreover, unlike HflX, the binding of HflXr induces conformational changes within adjacent rRNA nucleotides that would be incompatible with drug binding. These findings suggest that HflXr confers resistance using an allosteric ribosome protection mechanism, rather than by simply splitting and recycling antibiotic-stalled ribosomes.