Summary information and primary citation
- PDB-id
- 8aas; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- DNA binding protein
- Method
- X-ray (3.2 Å)
- Summary
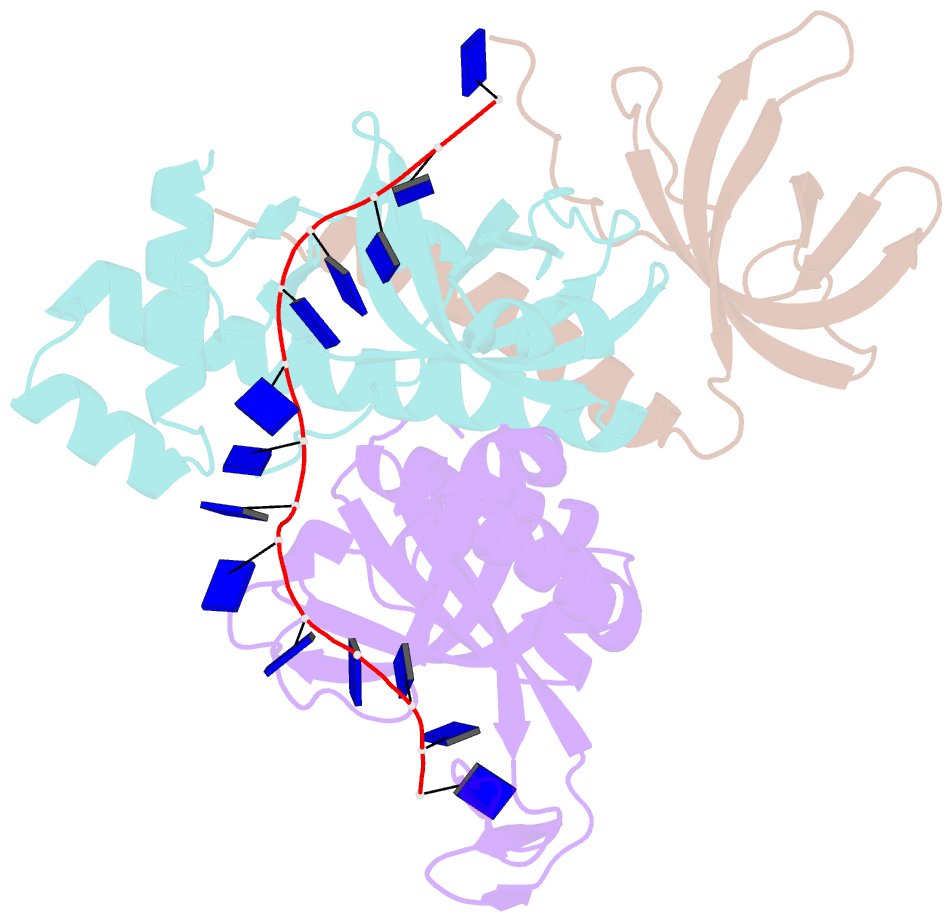
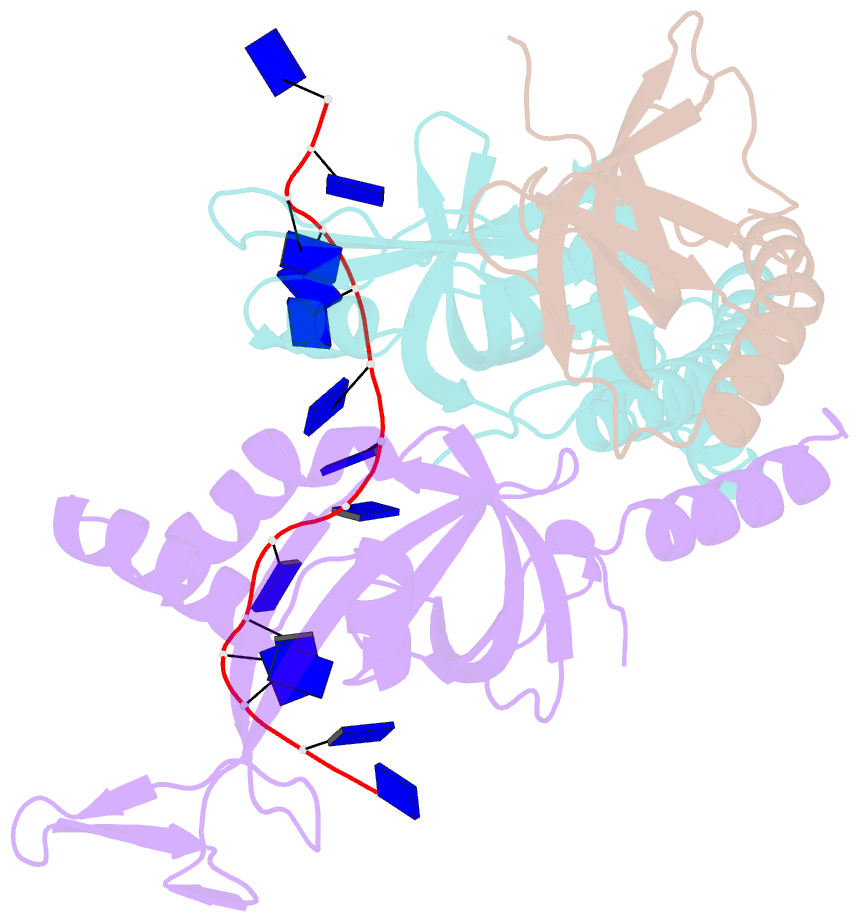
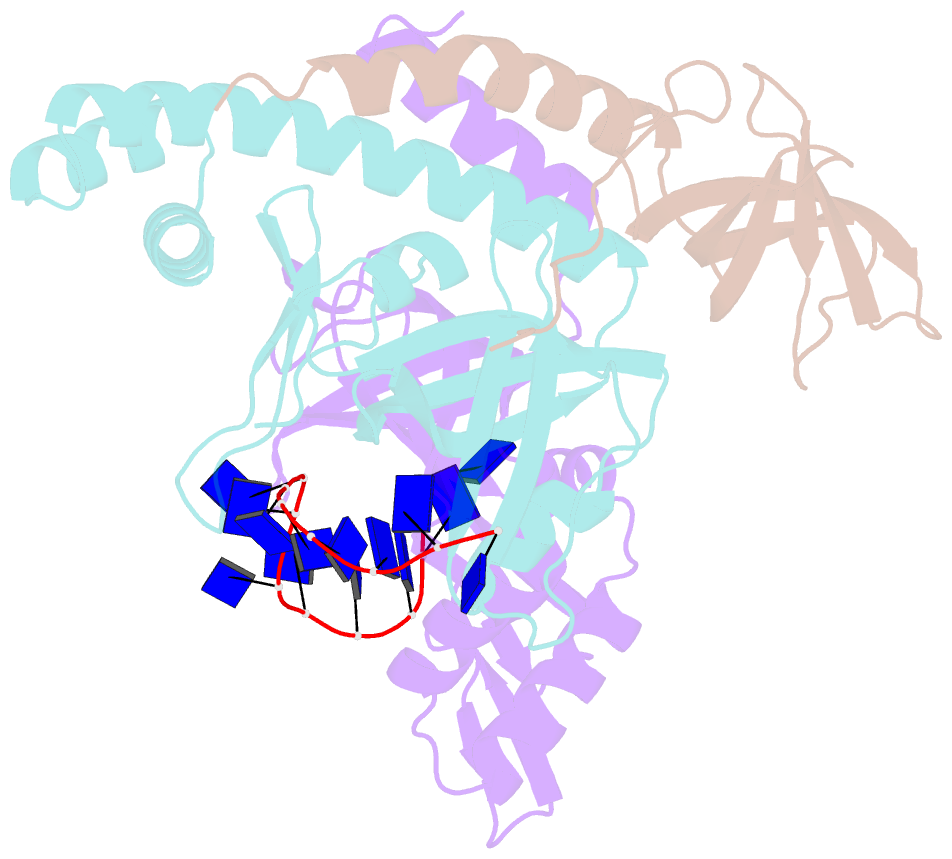
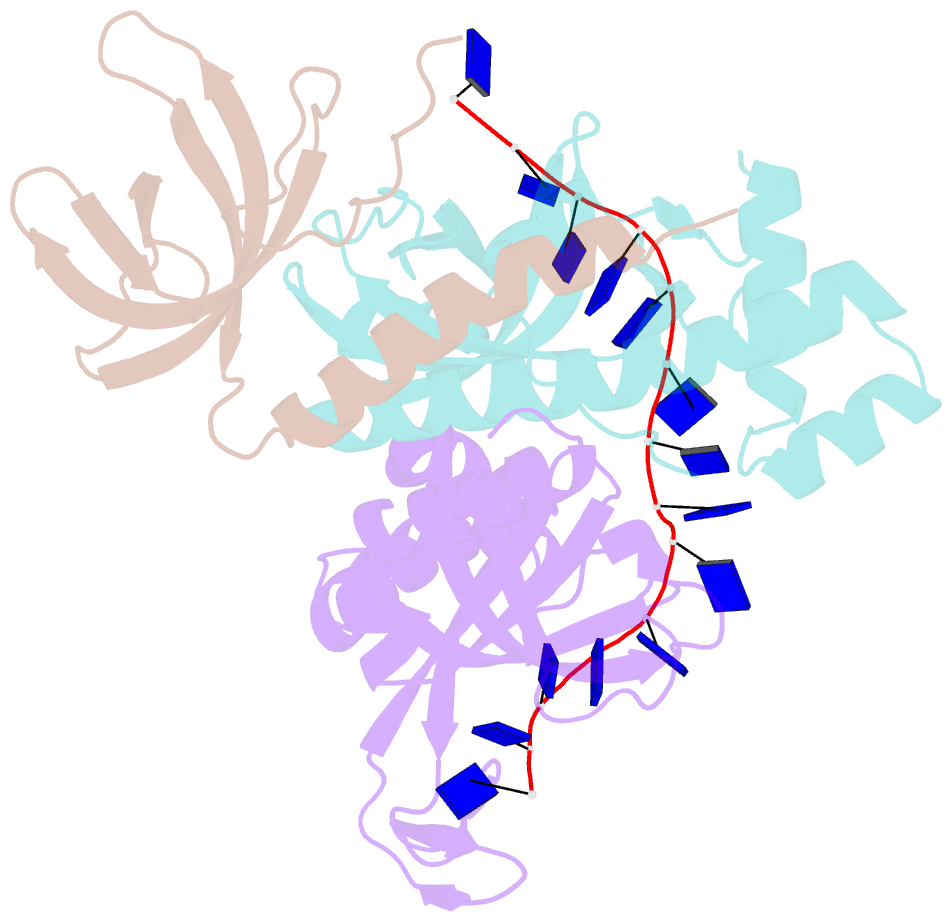
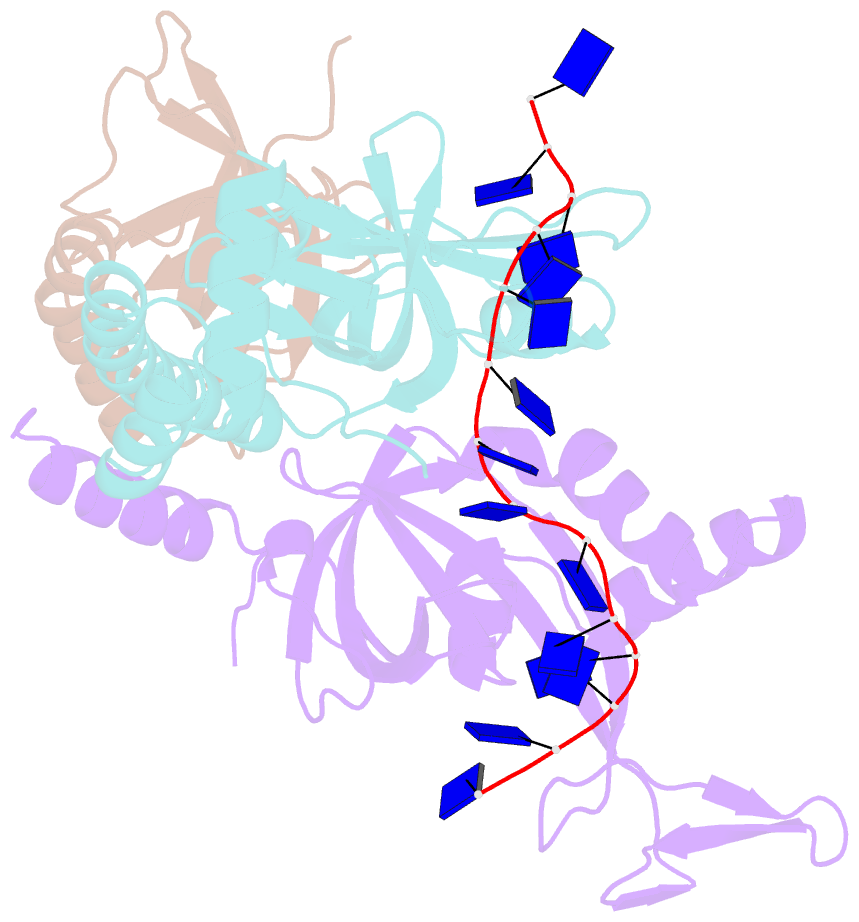
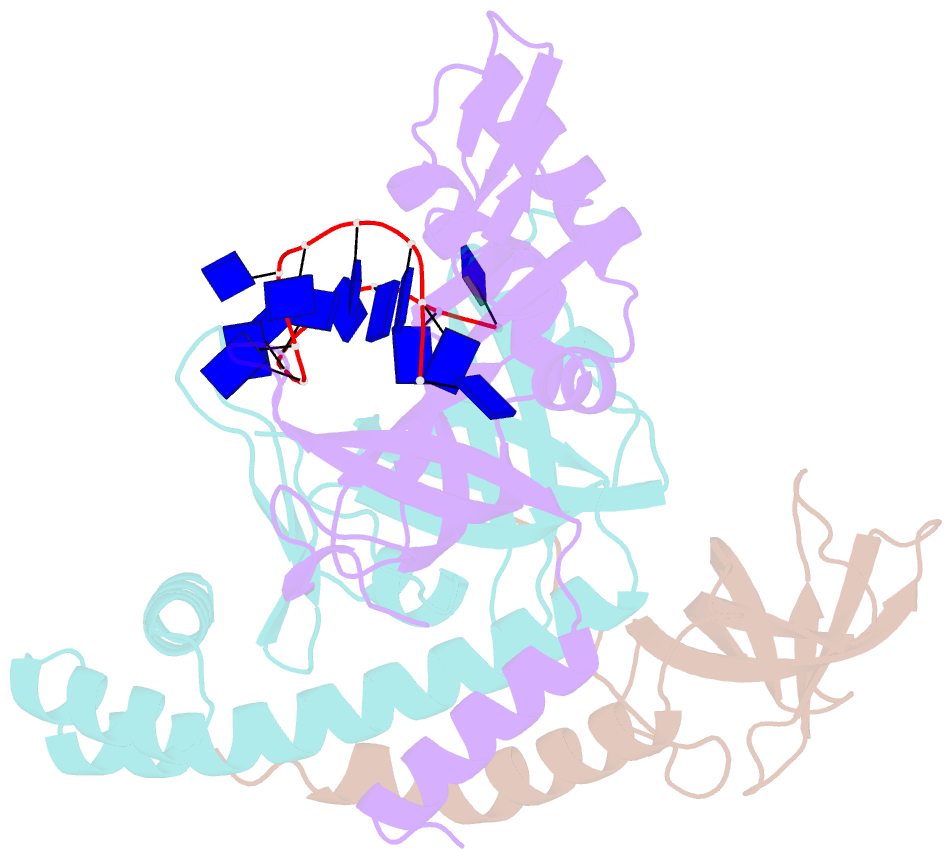
- Crystal structure of the pyrococcus abyssi rpa trimerization core bound to poly-dt20 ssDNA
- Reference
- Madru C, Martinez-Carranza M, Laurent S, Alberti AC, Chevreuil M, Raynal B, Haouz A, Le Meur RA, Delarue M, Henneke G, Flament D, Krupovic M, Legrand P, Sauguet L (2023): "DNA-binding mechanism and evolution of replication protein A." Nat Commun, 14, 2326. doi: 10.1038/s41467-023-38048-w.
- Abstract
- Replication Protein A (RPA) is a heterotrimeric single stranded DNA-binding protein with essential roles in DNA replication, recombination and repair. Little is known about the structure of RPA in Archaea, the third domain of life. By using an integrative structural, biochemical and biophysical approach, we extensively characterize RPA from Pyrococcus abyssi in the presence and absence of DNA. The obtained X-ray and cryo-EM structures reveal that the trimerization core and interactions promoting RPA clustering on ssDNA are shared between archaea and eukaryotes. However, we also identified a helical domain named AROD (Acidic Rpa1 OB-binding Domain), and showed that, in Archaea, RPA forms an unanticipated tetrameric supercomplex in the absence of DNA. The four RPA molecules clustered within the tetramer could efficiently coat and protect stretches of ssDNA created by the advancing replisome. Finally, our results provide insights into the evolution of this primordial replication factor in eukaryotes.