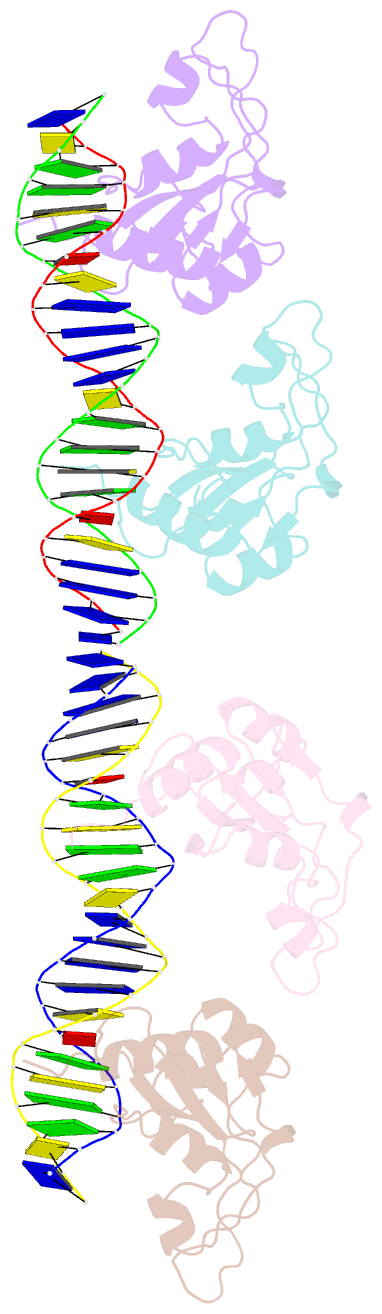
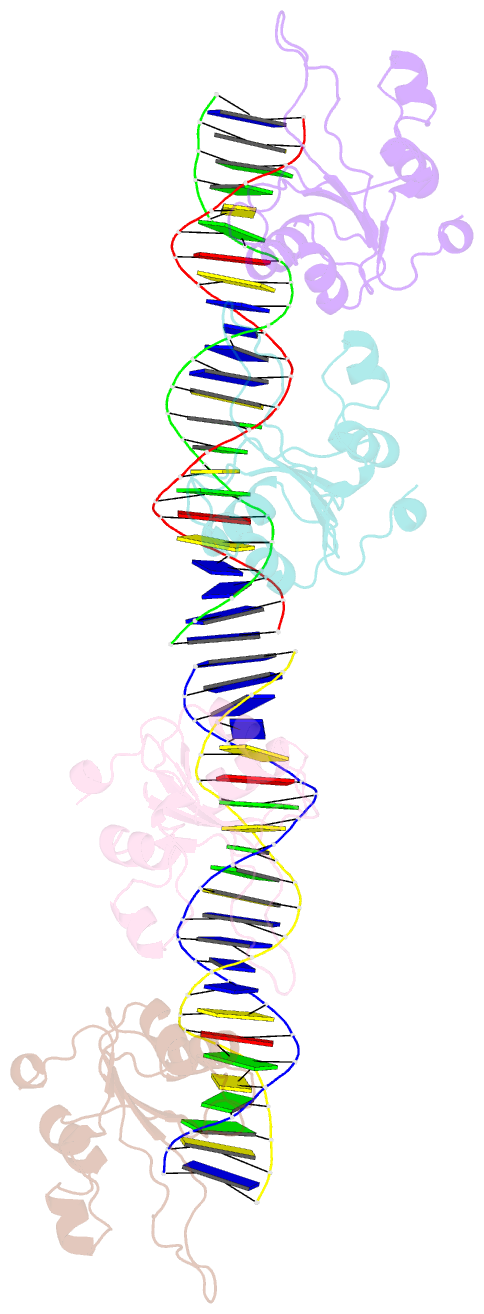
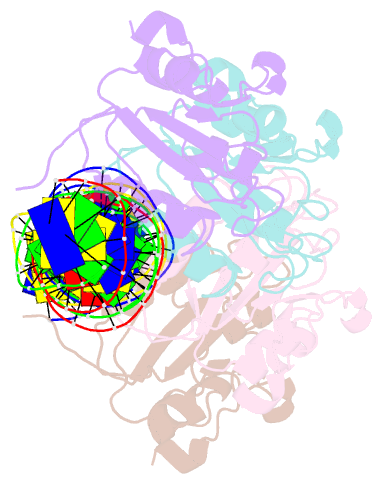
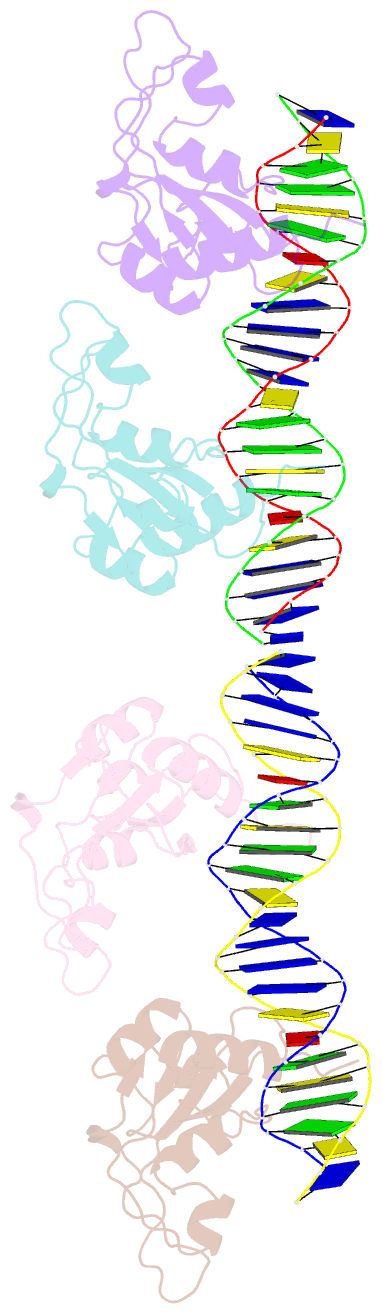
Summary information and primary citation
- PDB-id
- 8amu; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- replication
- Method
- X-ray (3.0 Å)
- Summary
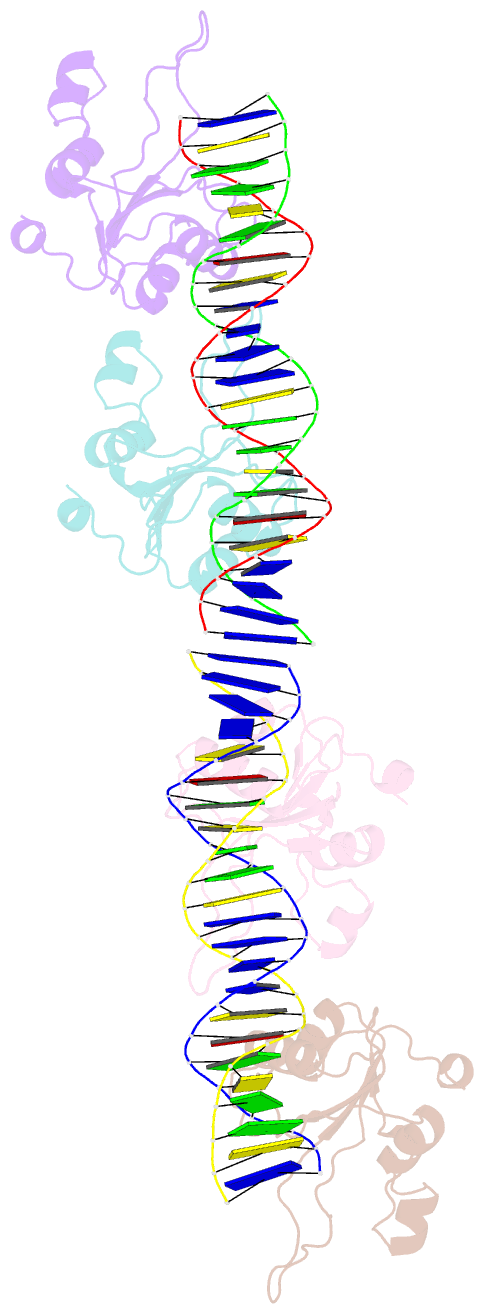
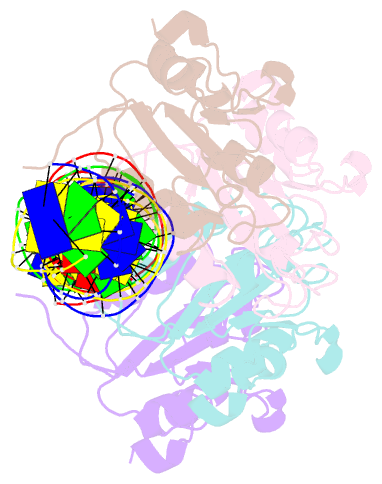
- Repb pmv158 obd domain bound to ddr region
- Reference
- Machon C, Ruiz-Maso JA, Amodio J, Boer DR, Bordanaba-Ruiseco L, Bury K, Konieczny I, Del Solar G, Coll M (2023): "Structures of pMV158 replication initiator RepB with and without DNA reveal a flexible dual-function protein." Nucleic Acids Res., 51, 1458-1472. doi: 10.1093/nar/gkac1271.
- Abstract
- DNA replication is essential to all living organisms as it ensures the fidelity of genetic material for the next generation of dividing cells. One of the simplest replication initiation mechanisms is the rolling circle replication. In the streptococcal plasmid pMV158, which confers antibiotic resistance to tetracycline, replication initiation is catalysed by RepB protein. The RepB N-terminal domain or origin binding domain binds to the recognition sequence (bind locus) of the double-strand origin of replication and cleaves one DNA strand at a specific site within the nic locus. Using biochemical and crystallographic analyses, here we show how the origin binding domain recognises and binds to the bind locus using structural elements removed from the active site, namely the recognition α helix, and a β-strand that organises upon binding. A new hexameric structure of full-length RepB that highlights the great flexibility of this protein is presented, which could account for its ability to perform different tasks, namely bind to two distinct loci and cleave one strand of DNA at the plasmid origin.