Summary information and primary citation
- PDB-id
- 8ane; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- immune system
- Method
- cryo-EM (3.2 Å)
- Summary
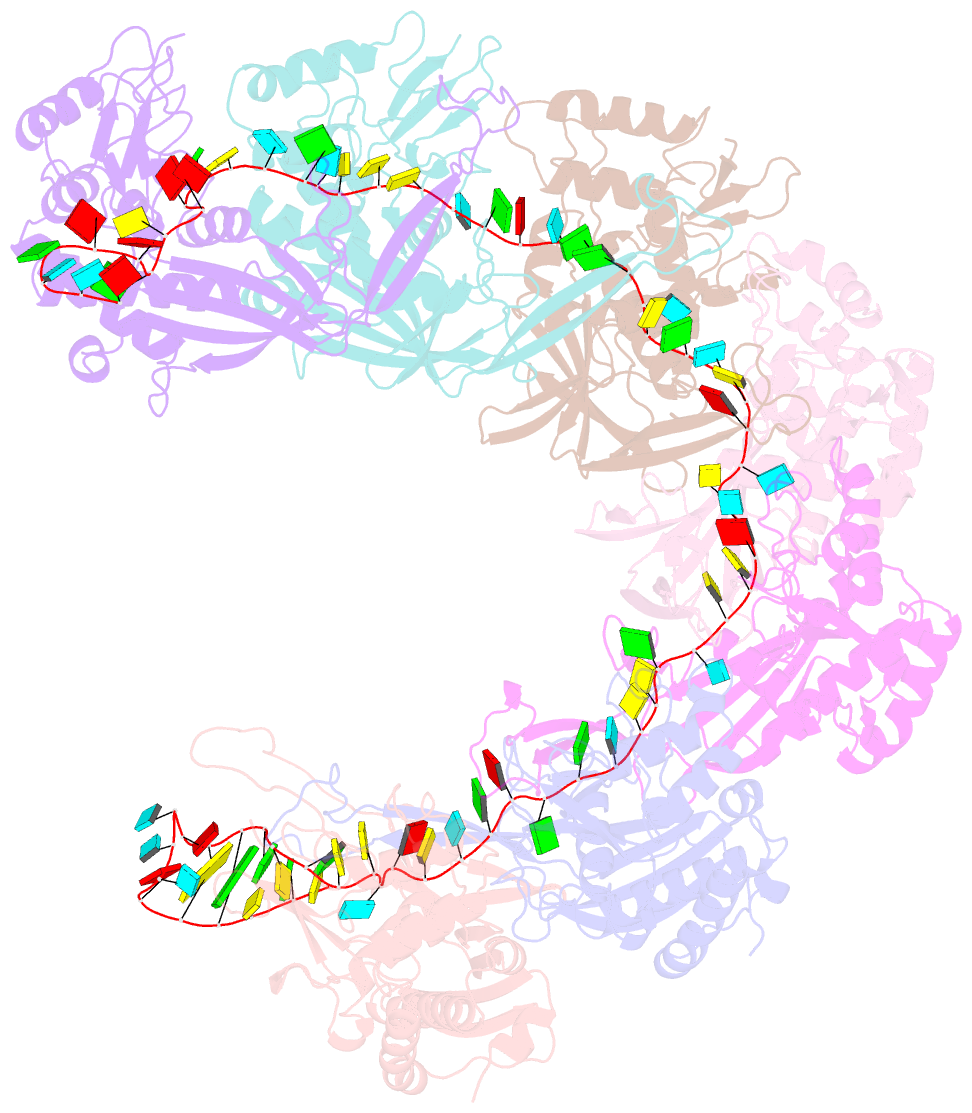
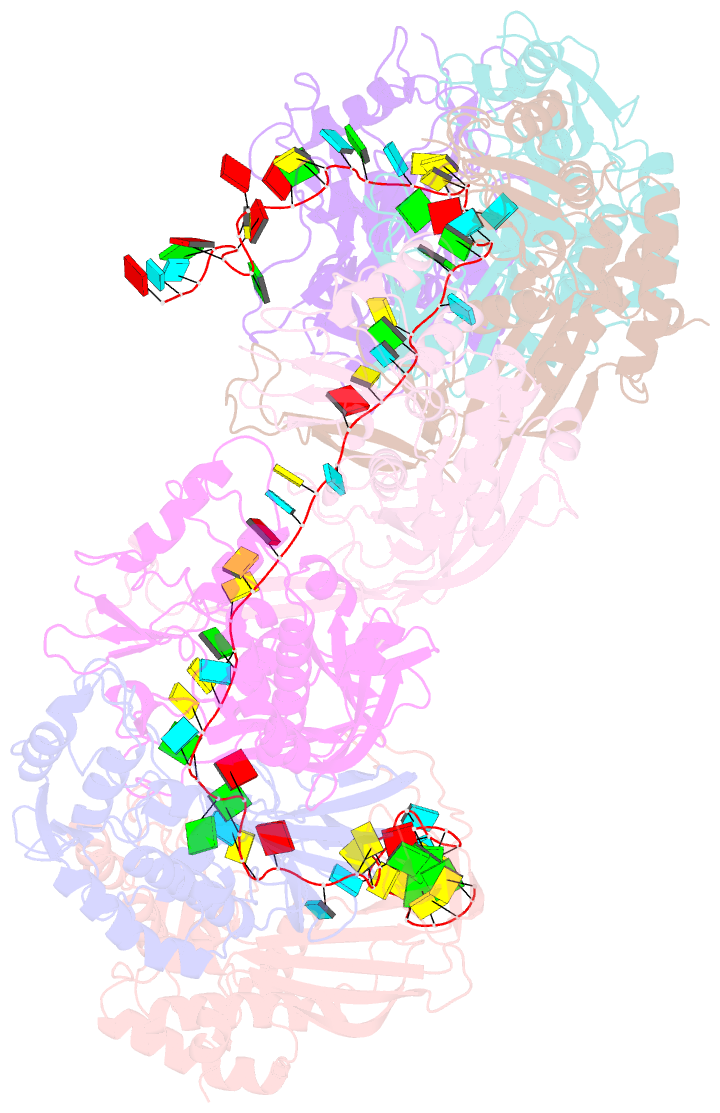
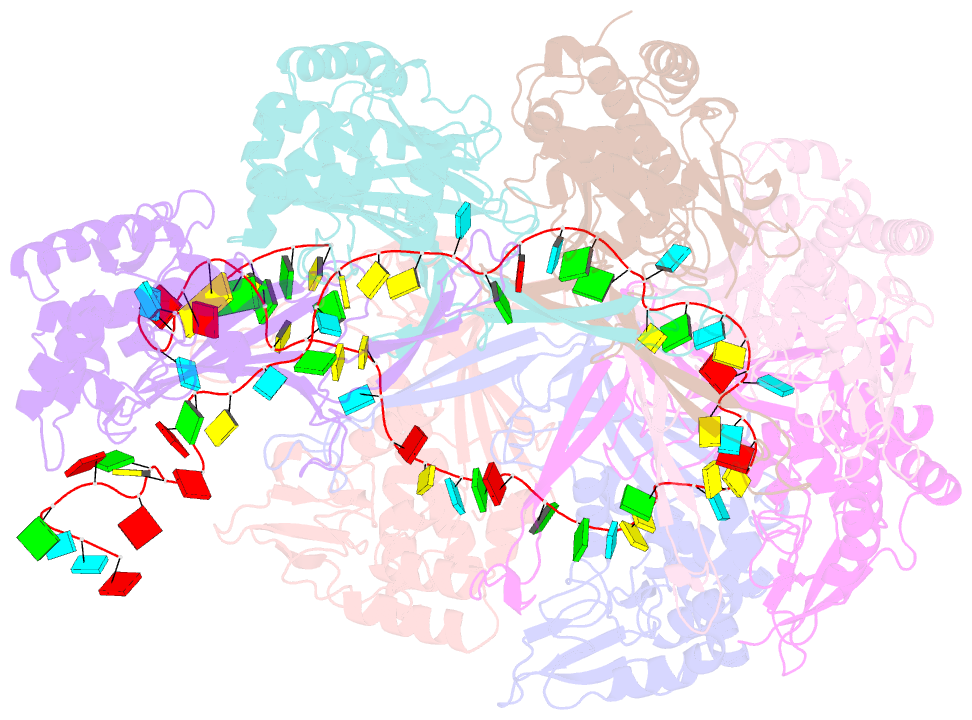
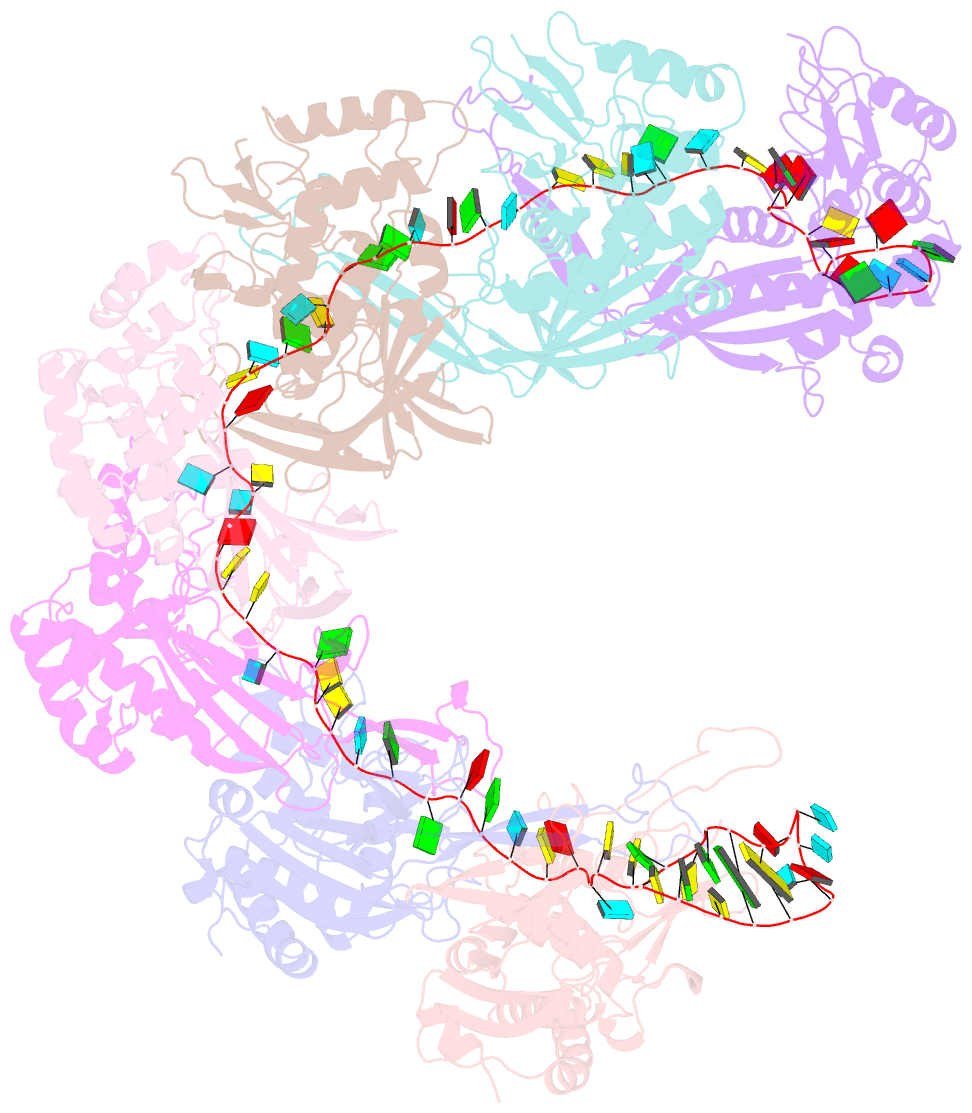
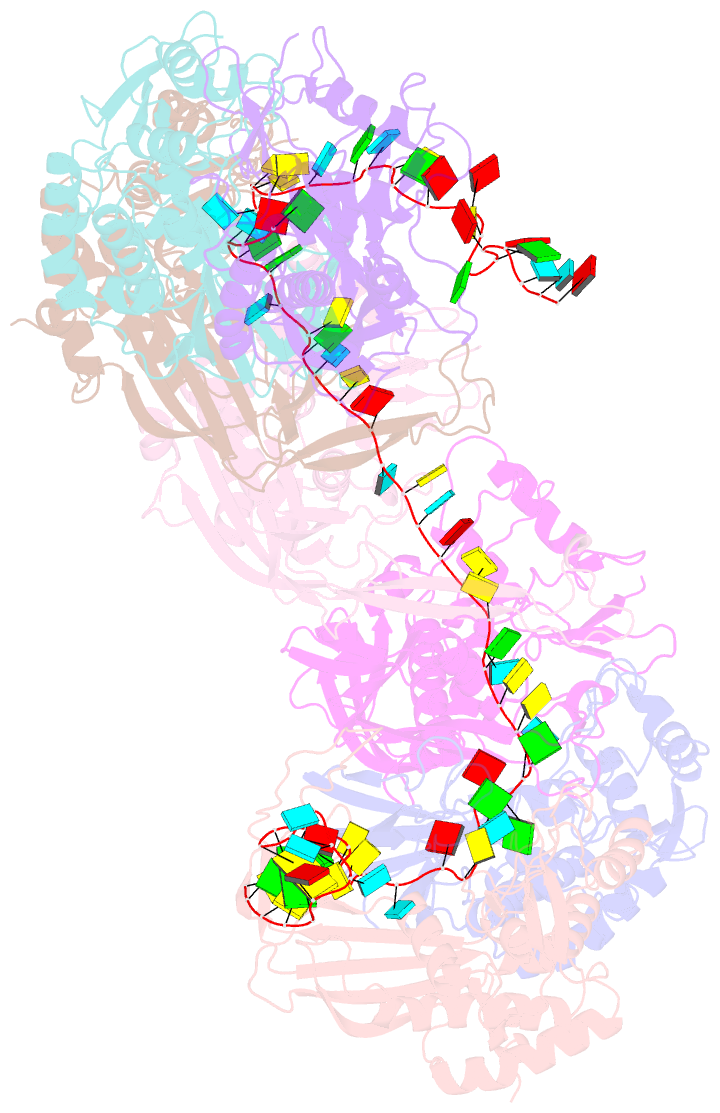
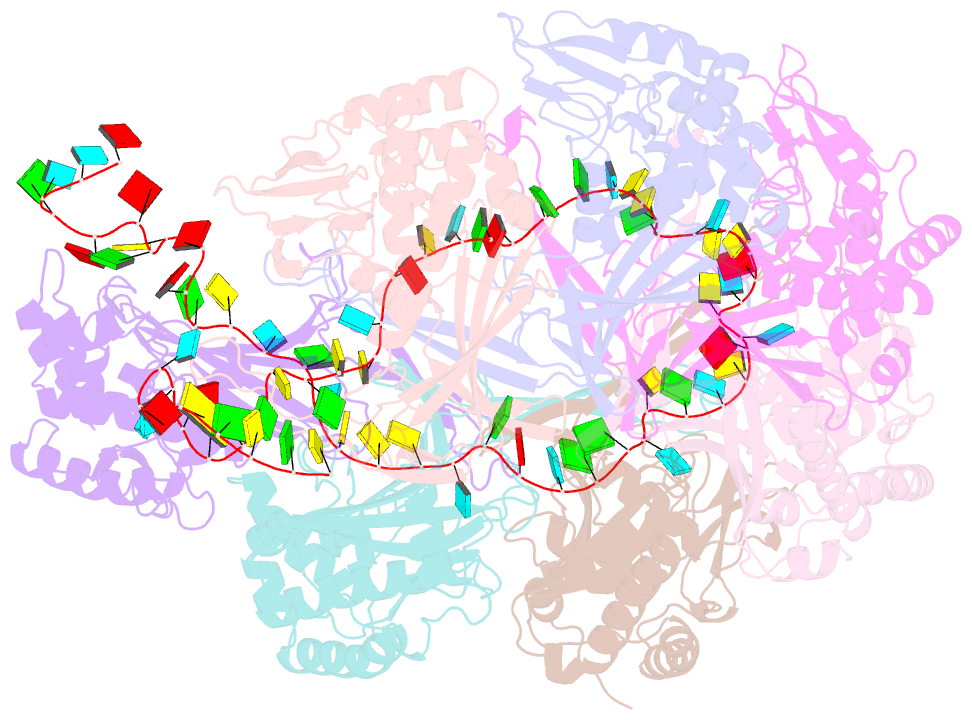
- Structure of the type i-g crispr effector
- Reference
- Shangguan Q, Graham S, Sundaramoorthy R, White MF (2022): "Structure and mechanism of the type I-G CRISPR effector." Nucleic Acids Res., 50, 11214-11228. doi: 10.1093/nar/gkac925.
- Abstract
- Type I CRISPR systems are the most common CRISPR type found in bacteria. They use a multisubunit effector, guided by crRNA, to detect and bind dsDNA targets, forming an R-loop and recruiting the Cas3 enzyme to facilitate target DNA destruction, thus providing immunity against mobile genetic elements. Subtypes have been classified into families A-G, with type I-G being the least well understood. Here, we report the composition, structure and function of the type I-G Cascade CRISPR effector from Thioalkalivibrio sulfidiphilus, revealing key new molecular details. The unique Csb2 subunit processes pre-crRNA, remaining bound to the 3' end of the mature crRNA, and seven Cas7 subunits form the backbone of the effector. Cas3 associates stably with the effector complex via the Cas8g subunit and is important for target DNA recognition. Structural analysis by cryo-Electron Microscopy reveals a strikingly curved backbone conformation with Cas8g spanning the belly of the structure. These biochemical and structural insights shed new light on the diversity of type I systems and open the way to applications in genome engineering.