Summary information and primary citation
- PDB-id
- 8apm; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- viral protein
- Method
- cryo-EM (6.6 Å)
- Summary
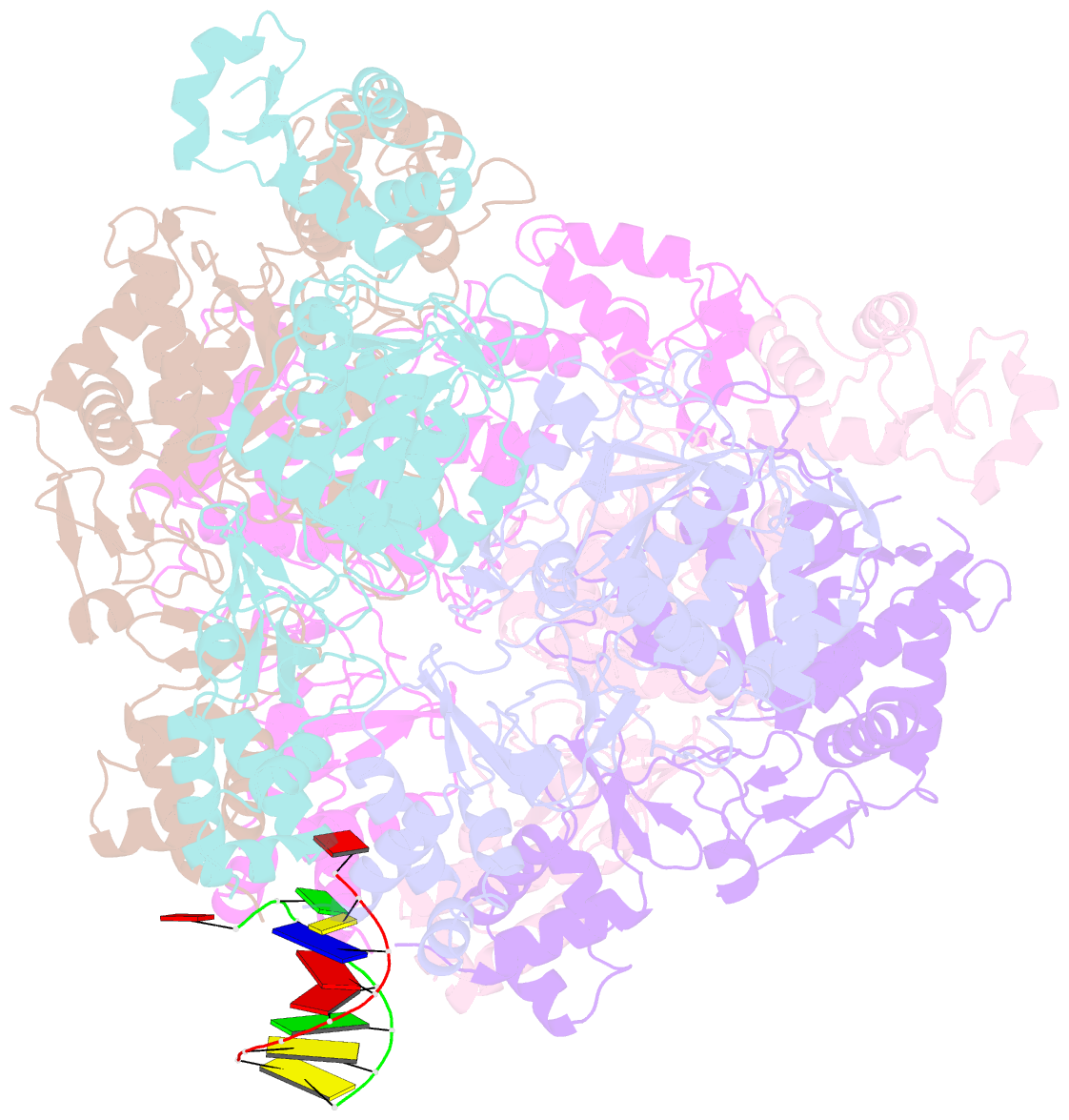
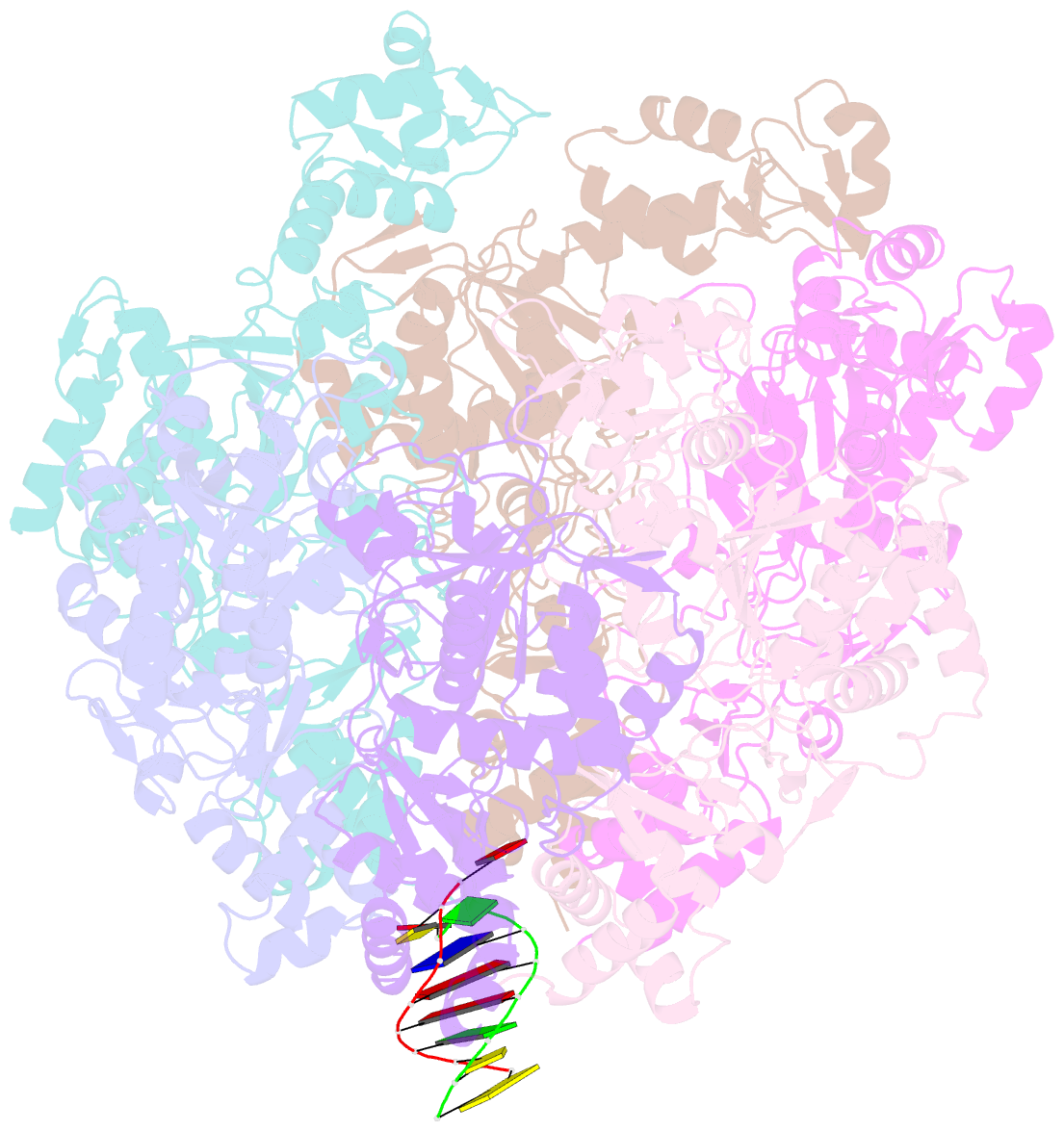
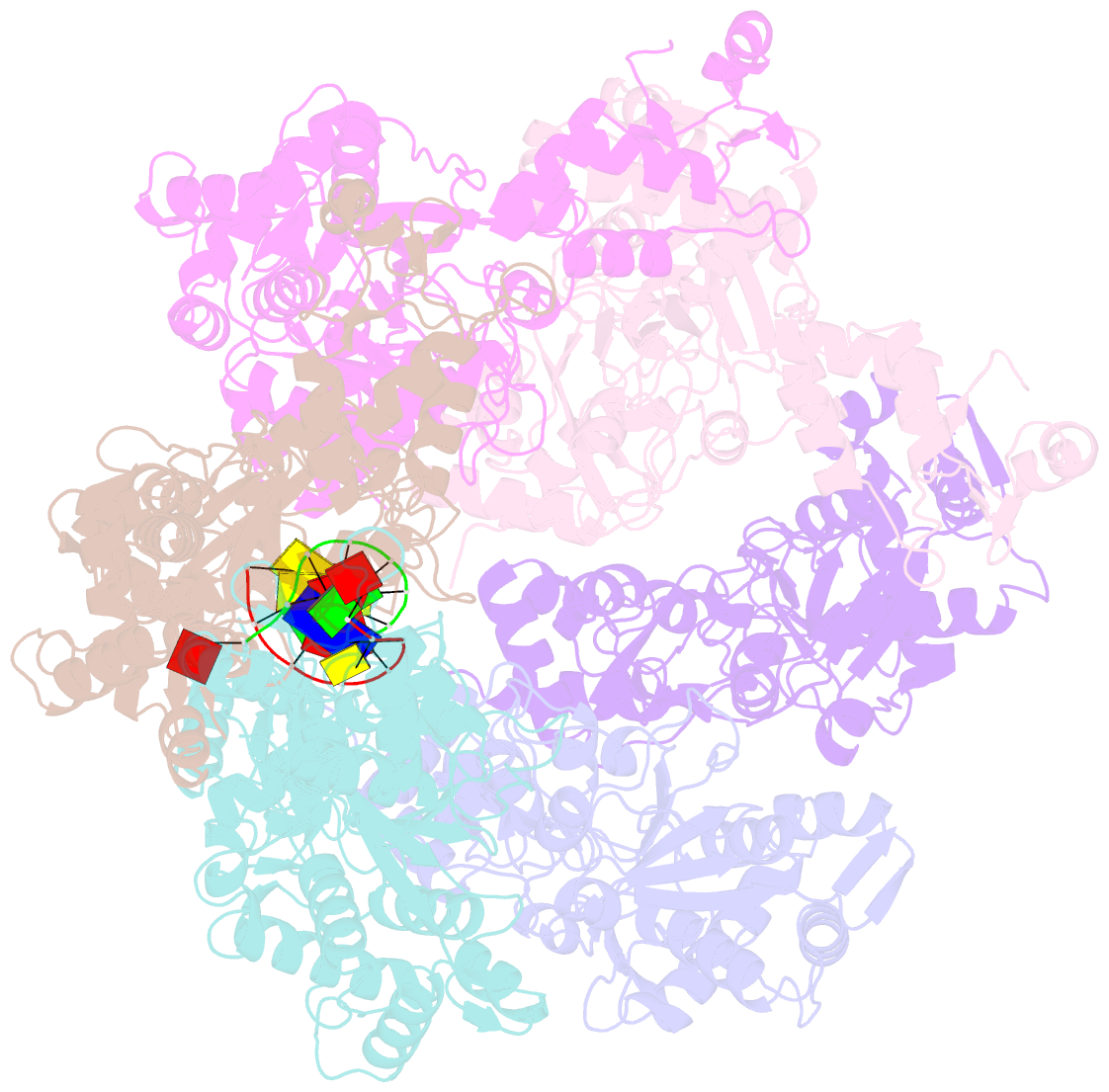
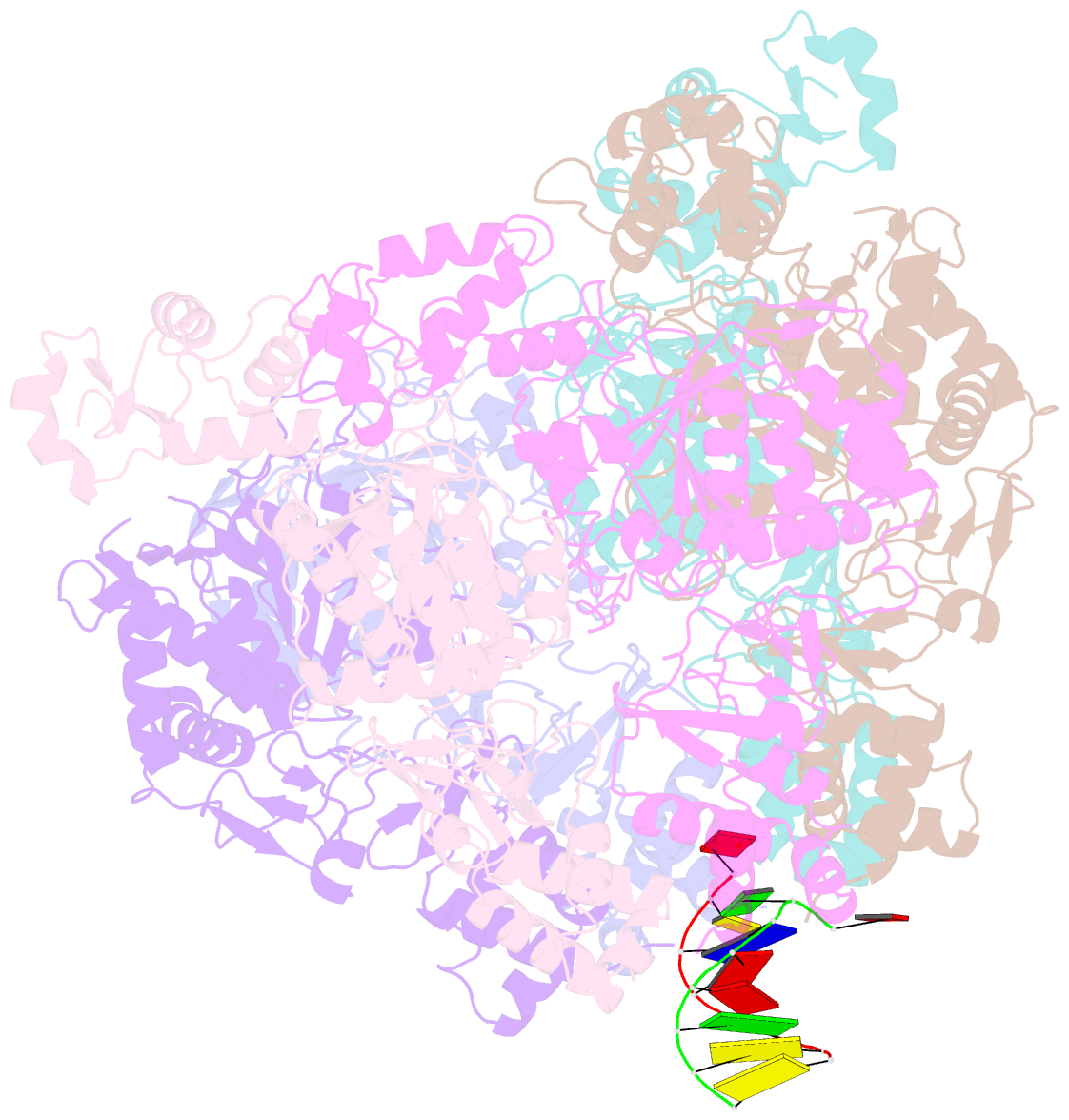
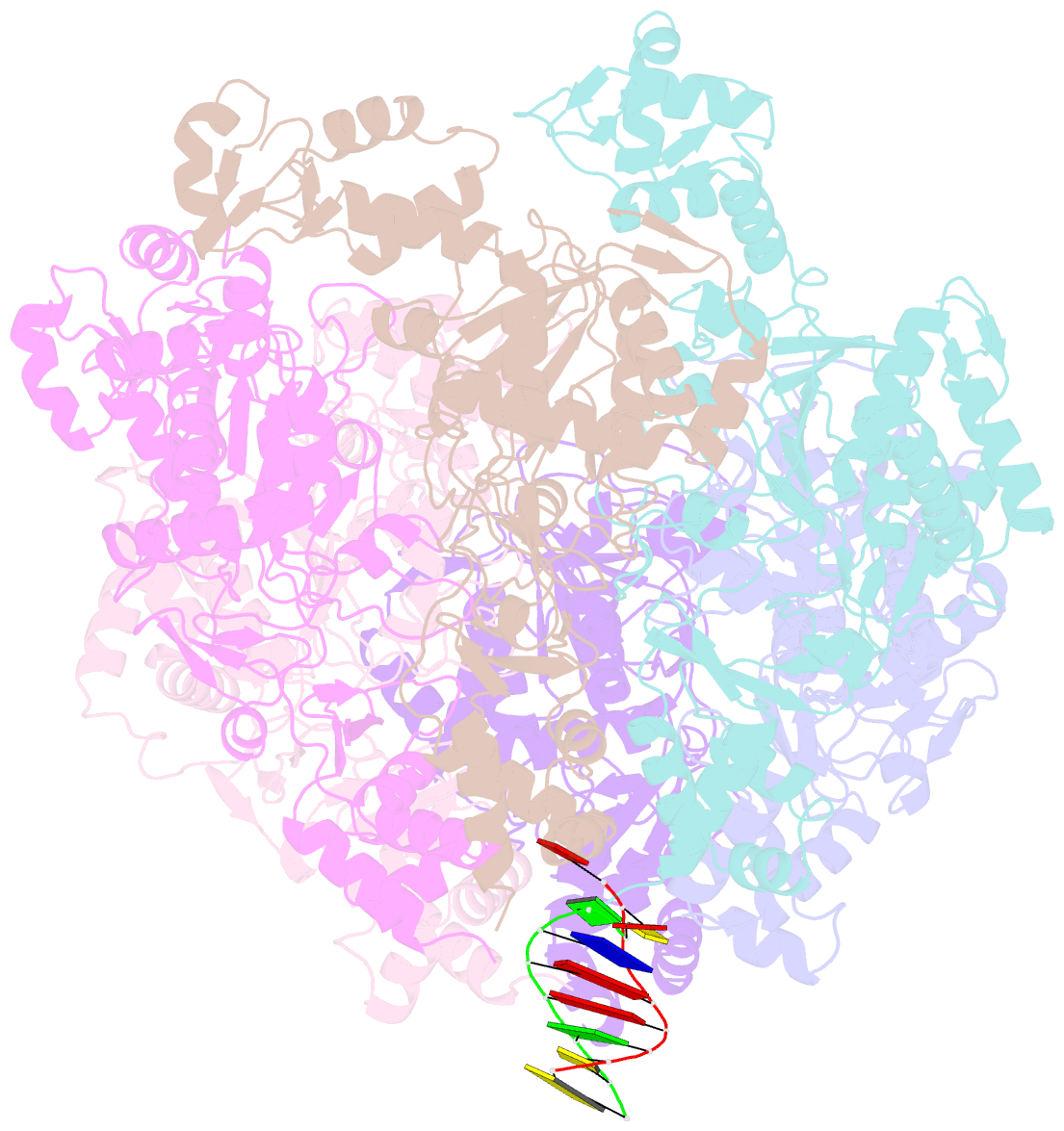
- Vaccinia virus DNA helicase d5 residues 323-785 hexamer with bound DNA processed in c1
- Reference
- Hutin S, Ling WL, Tarbouriech N, Schoehn G, Grimm C, Fischer U, Burmeister WP (2022): "The Vaccinia Virus DNA Helicase Structure from Combined Single-Particle Cryo-Electron Microscopy and AlphaFold2 Prediction." Viruses, 14. doi: 10.3390/v14102206.
- Abstract
- Poxviruses are large DNA viruses with a linear double-stranded DNA genome circularized at the extremities. The helicase-primase D5, composed of six identical 90 kDa subunits, is required for DNA replication. D5 consists of a primase fragment flexibly attached to the hexameric C-terminal polypeptide (res. 323-785) with confirmed nucleotide hydrolase and DNA-binding activity but an elusive helicase activity. We determined its structure by single-particle cryo-electron microscopy. It displays an AAA+ helicase core flanked by N- and C-terminal domains. Model building was greatly helped by the predicted structure of D5 using AlphaFold2. The 3.9 Å structure of the N-terminal domain forms a well-defined tight ring while the resolution decreases towards the C-terminus, still allowing the fit of the predicted structure. The N-terminal domain is partially present in papillomavirus E1 and polyomavirus LTA helicases, as well as in a bacteriophage NrS-1 helicase domain, which is also closely related to the AAA+ helicase domain of D5. Using the Pfam domain database, a D5_N domain followed by DUF5906 and Pox_D5 domains could be assigned to the cryo-EM structure, providing the first 3D structures for D5_N and Pox_D5 domains. The same domain organization has been identified in a family of putative helicases from large DNA viruses, bacteriophages, and selfish DNA elements.