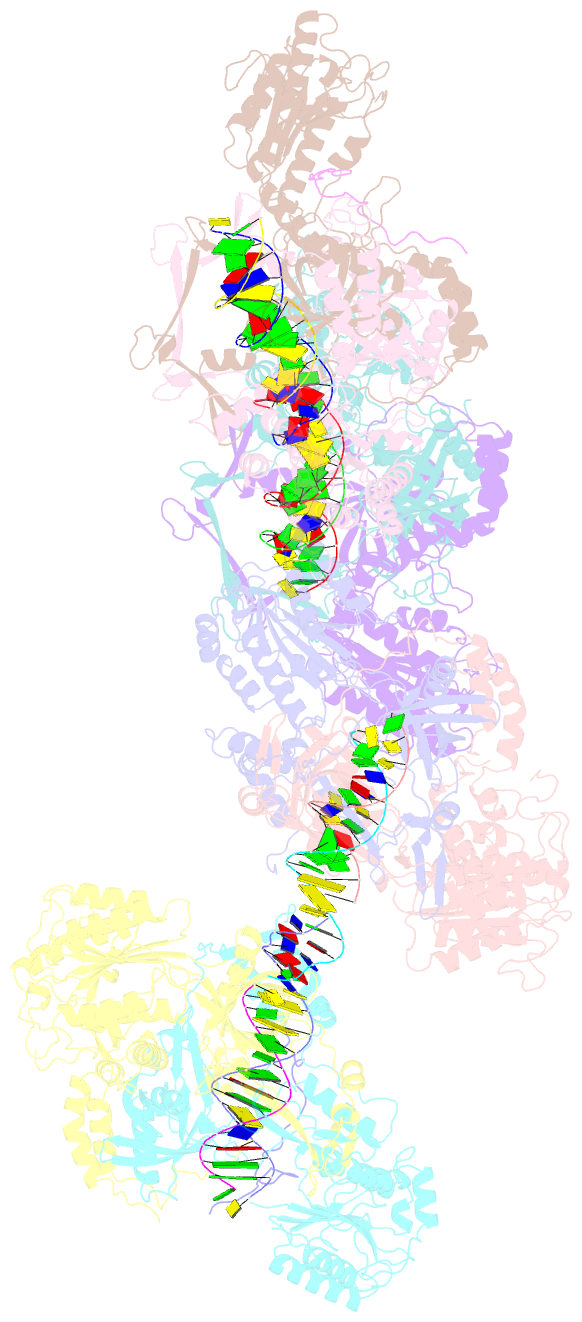
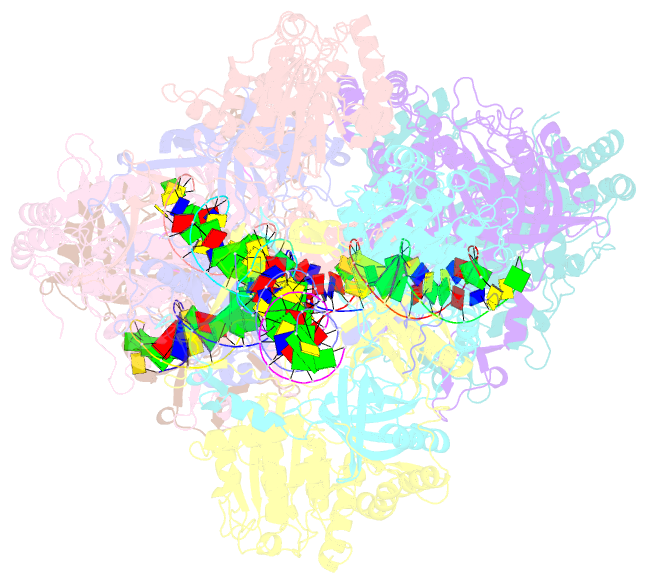
Summary information and primary citation
- PDB-id
- 8asc; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- DNA binding protein
- Method
- X-ray (2.95 Å)
- Summary
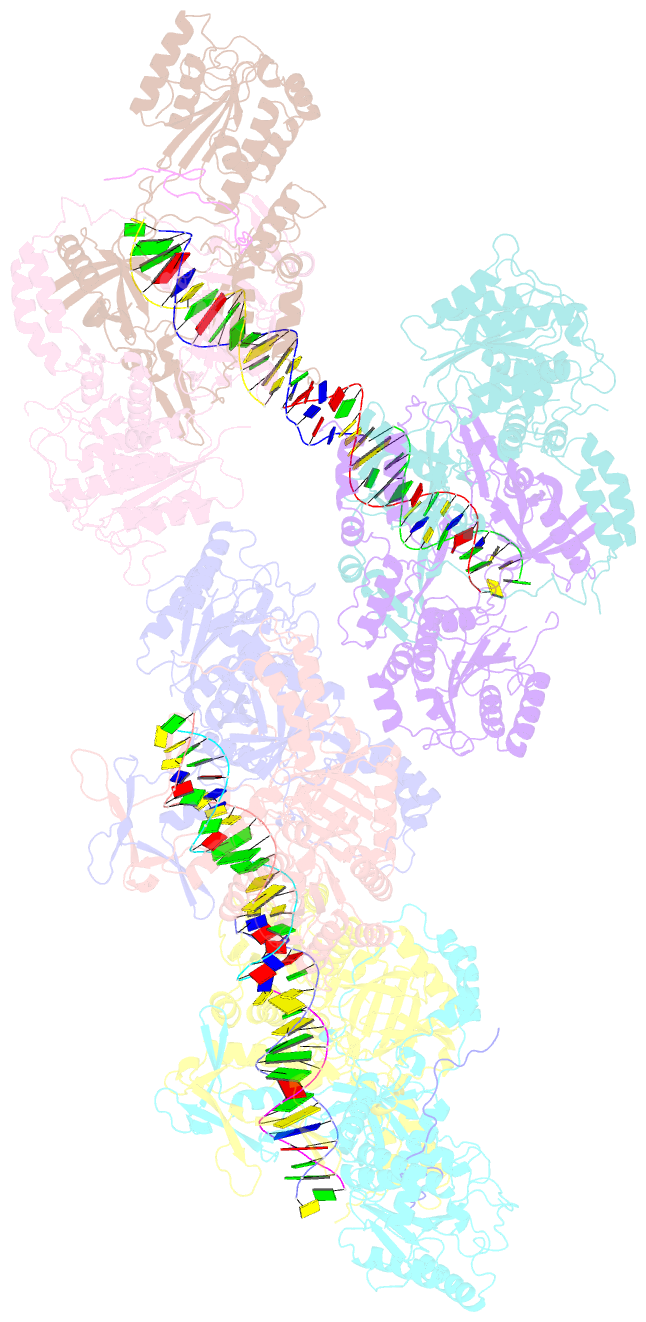
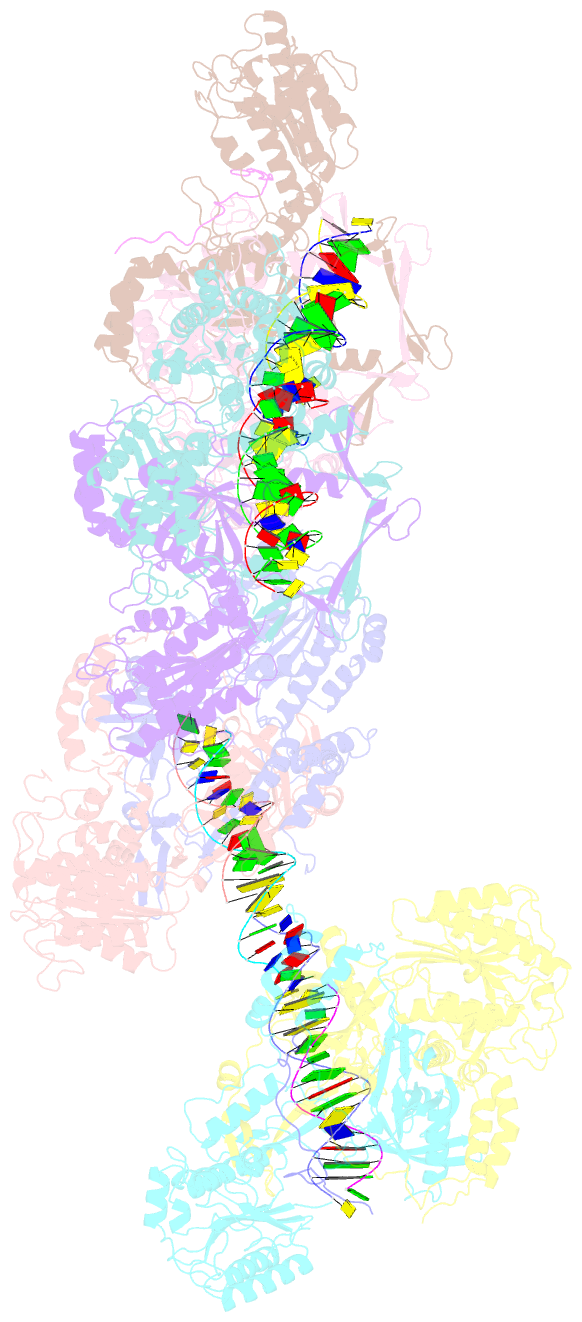
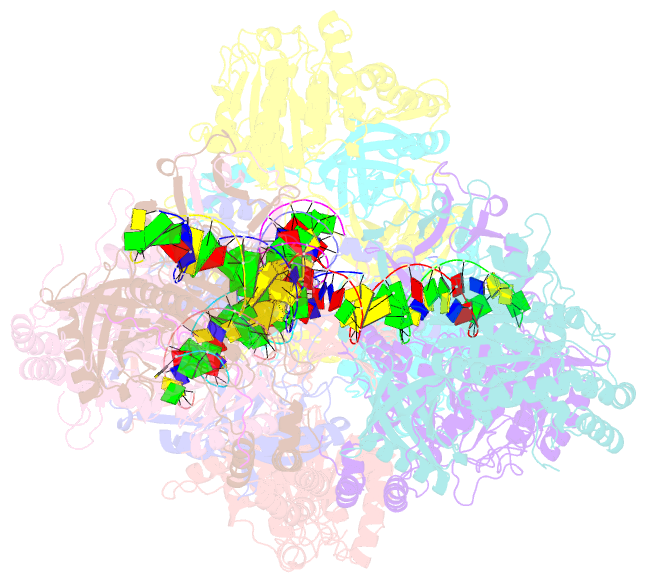
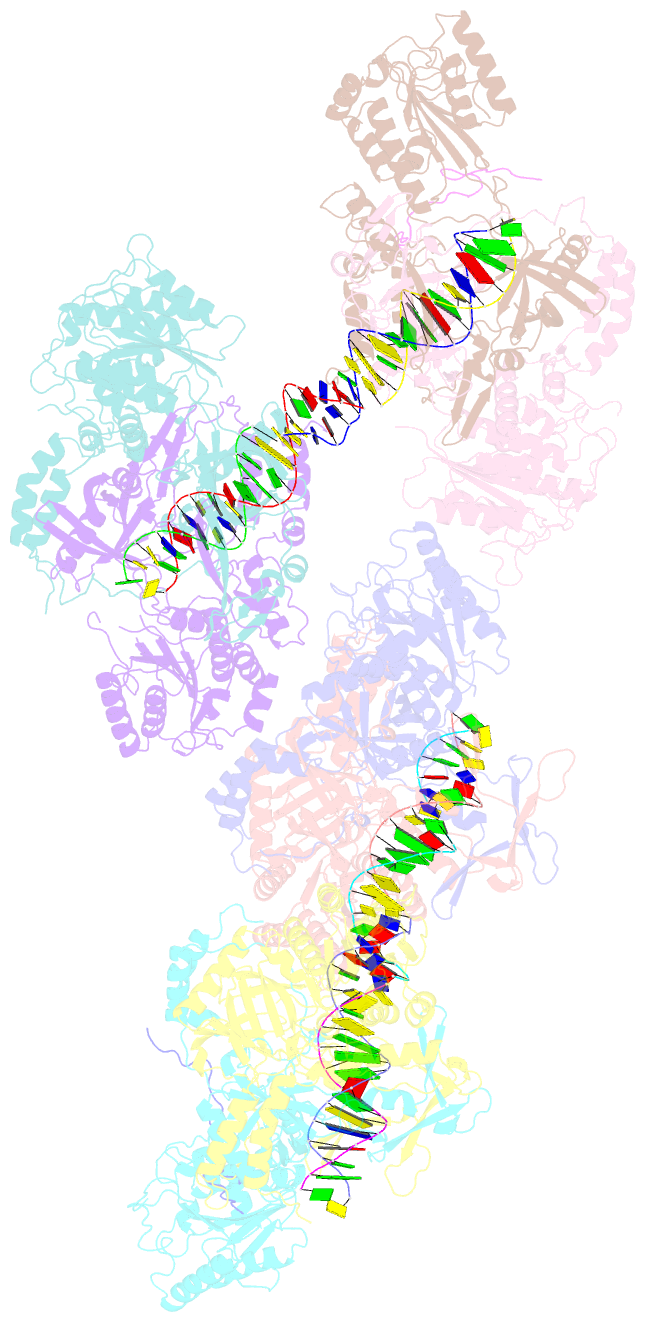
- Ku70-80 binds to the ku-binding motif of paxx
- Reference
- Seif-El-Dahan M, Kefala-Stavridi A, Frit P, Hardwick SW, Chirgadze DY, Maia De Oliviera T, Britton S, Barboule N, Bossaert M, Pandurangan AP, Meek K, Blundell TL, Ropars V, Calsou P, Charbonnier JB, Chaplin AK (2023): "PAXX binding to the NHEJ machinery explains functional redundancy with XLF." Sci Adv, 9, eadg2834. doi: 10.1126/sciadv.adg2834.
- Abstract
- Nonhomologous end joining is a critical mechanism that repairs DNA double-strand breaks in human cells. In this work, we address the structural and functional role of the accessory protein PAXX [paralog of x-ray repair cross-complementing protein 4 (XRCC4) and XRCC4-like factor (XLF)] in this mechanism. Here, we report high-resolution cryo-electron microscopy (cryo-EM) and x-ray crystallography structures of the PAXX C-terminal Ku-binding motif bound to Ku70/80 and cryo-EM structures of PAXX bound to two alternate DNA-dependent protein kinase (DNA-PK) end-bridging dimers, mediated by either Ku80 or XLF. We identify residues critical for the Ku70/PAXX interaction in vitro and in cells. We demonstrate that PAXX and XLF can bind simultaneously to the Ku heterodimer and act as structural bridges in alternate forms of DNA-PK dimers. Last, we show that engagement of both proteins provides a complementary advantage for DNA end synapsis and end joining in cells.