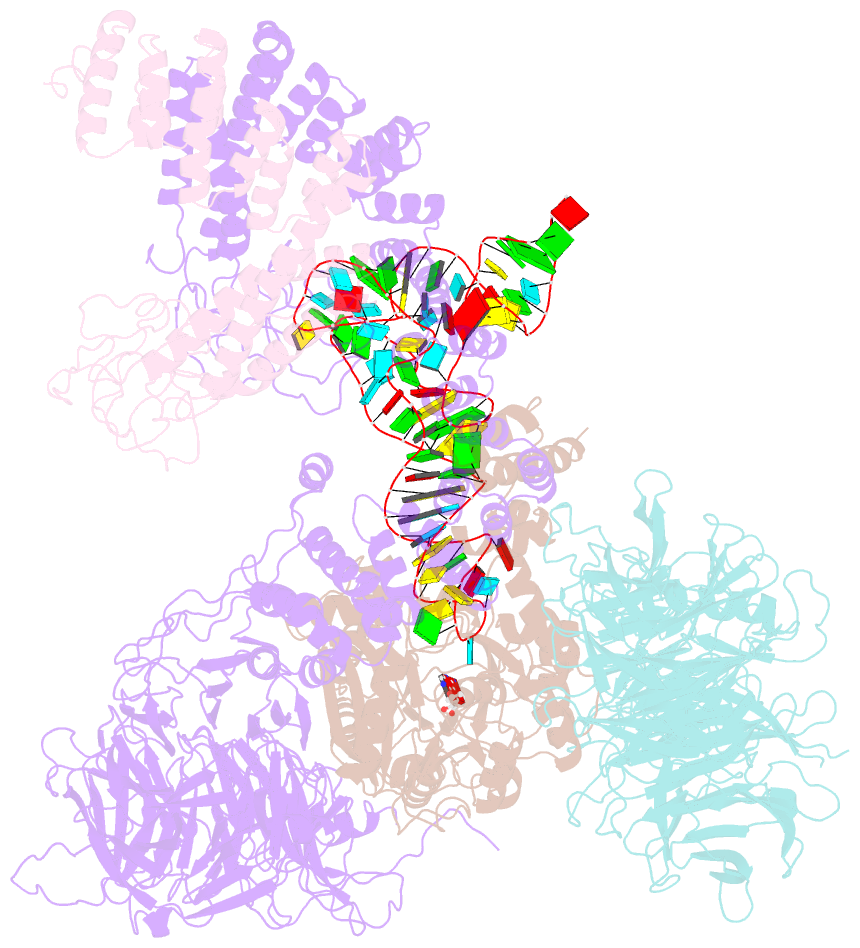
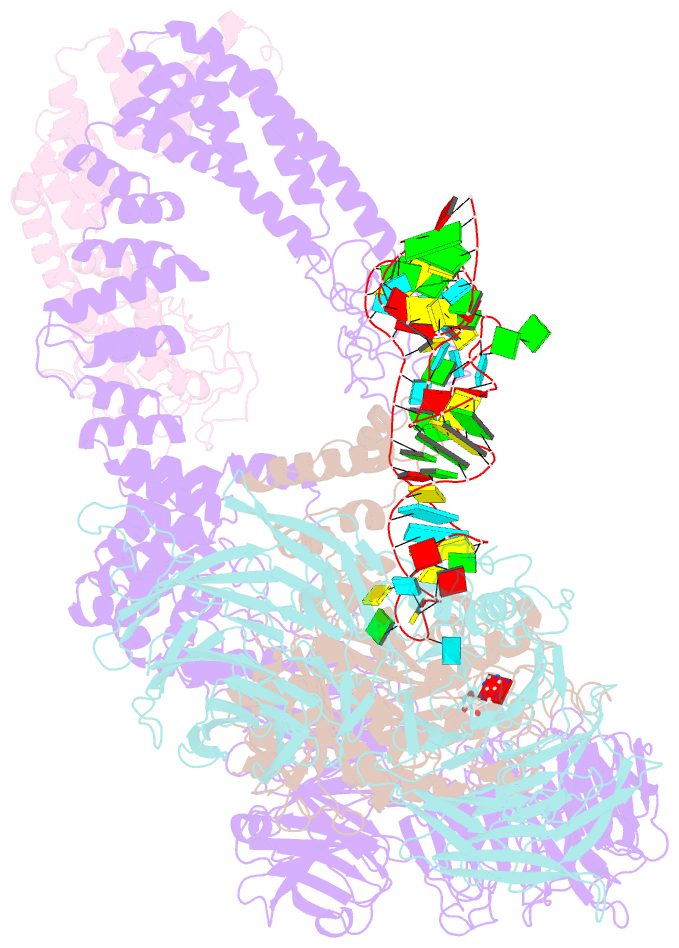
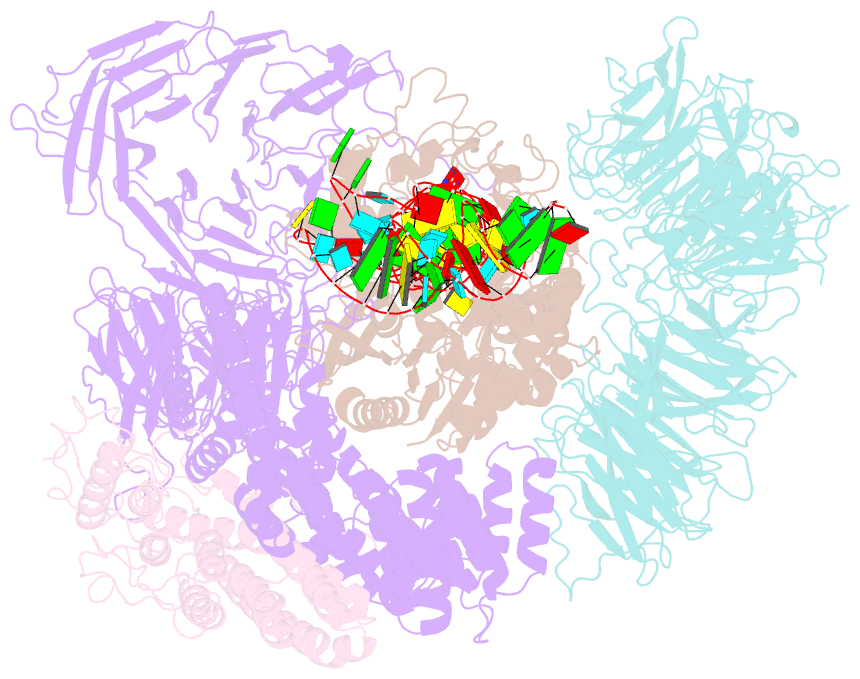
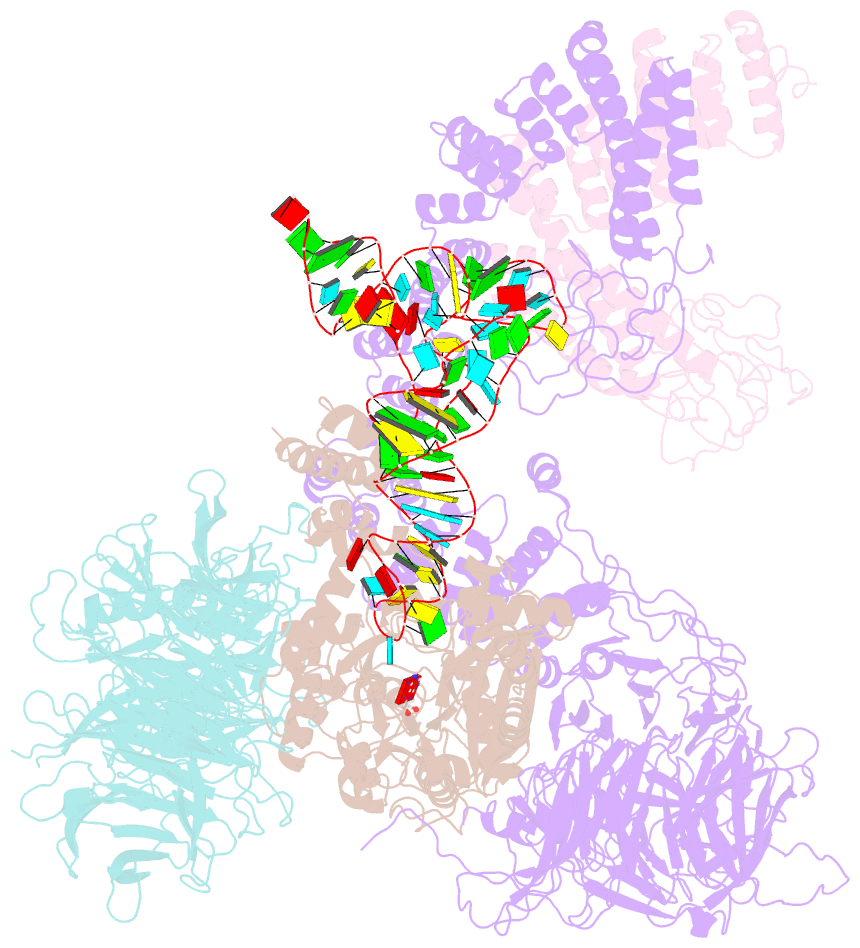
Summary information and primary citation
- PDB-id
- 8asw; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- translation
- Method
- cryo-EM (3.96 Å)
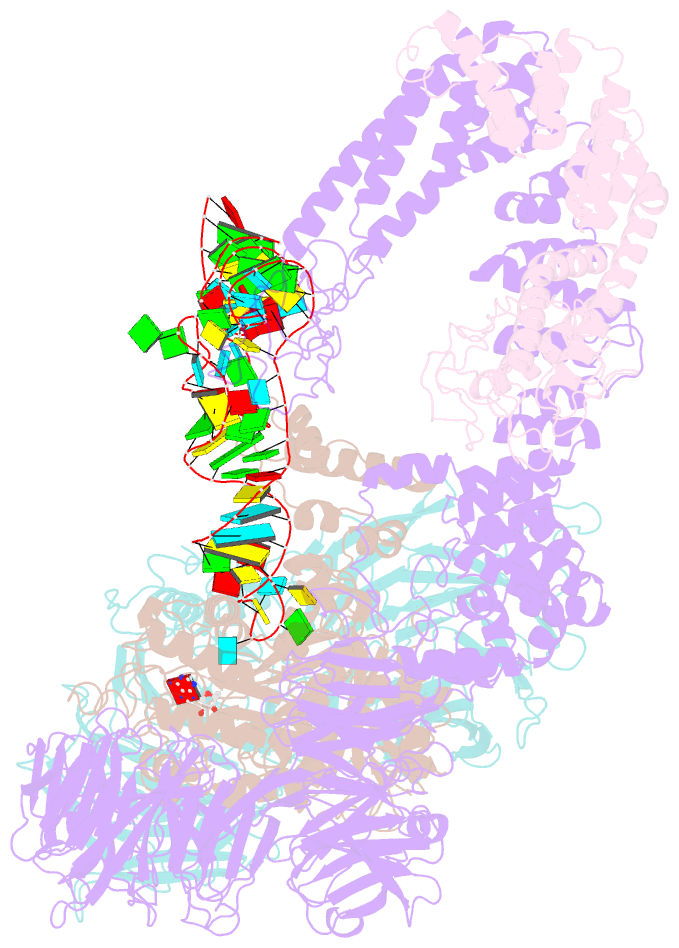
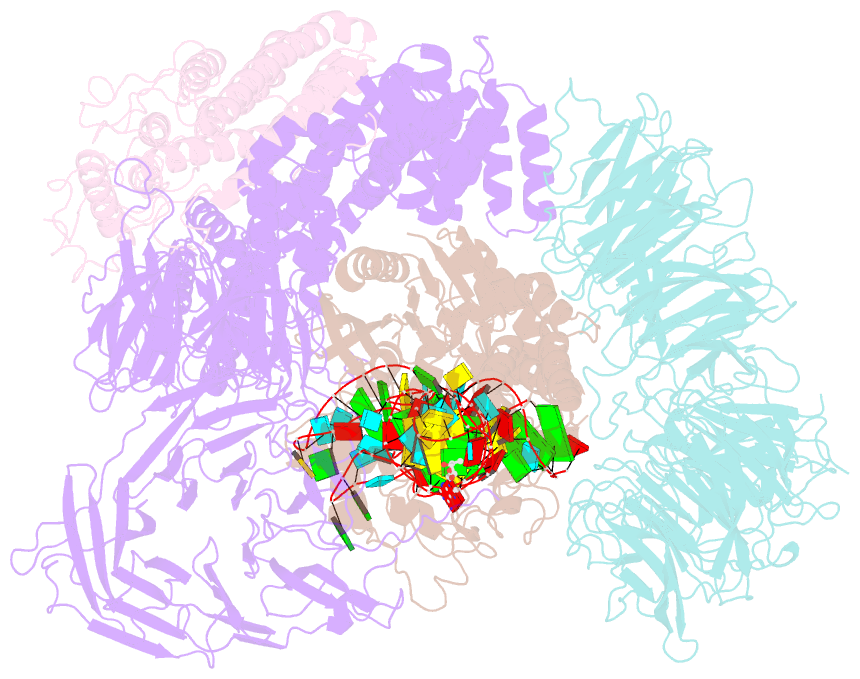
- Summary
- cryo-EM structure of yeast elp123 in complex with alanine trna
- Reference
- Jaciuk M, Scherf D, Kaszuba K, Gaik M, Rau A, Koscielniak A, Krutyholowa R, Rawski M, Indyka P, Graziadei A, Chramiec-Glabik A, Biela A, Dobosz D, Lin TY, Abbassi NE, Hammermeister A, Rappsilber J, Kosinski J, Schaffrath R, Glatt S (2023): "Cryo-EM structure of the fully assembled Elongator complex." Nucleic Acids Res., 51, 2011-2032. doi: 10.1093/nar/gkac1232.
- Abstract
- The multi-subunit Elongator complex mediates the addition of a carboxymethyl group to wobble uridines in eukaryotic tRNAs. This tRNA modification is crucial to preserve the integrity of cellular proteomes and to protects us against severe neurodegenerative diseases. Elongator is organized in two distinct modules (i) the larger Elp123 subcomplex that binds and modifies the suitable tRNA substrate and (ii) the smaller Elp456 subcomplex that assists the release of the modified tRNA. The presented cryo-EM structures of Elongator show that the assemblies are very dynamic and undergo conformational rearrangements at consecutive steps of the process. Last but not least, the study provides a detailed reaction scheme and shows that the architecture of Elongator is highly conserved from yeast to mammals.