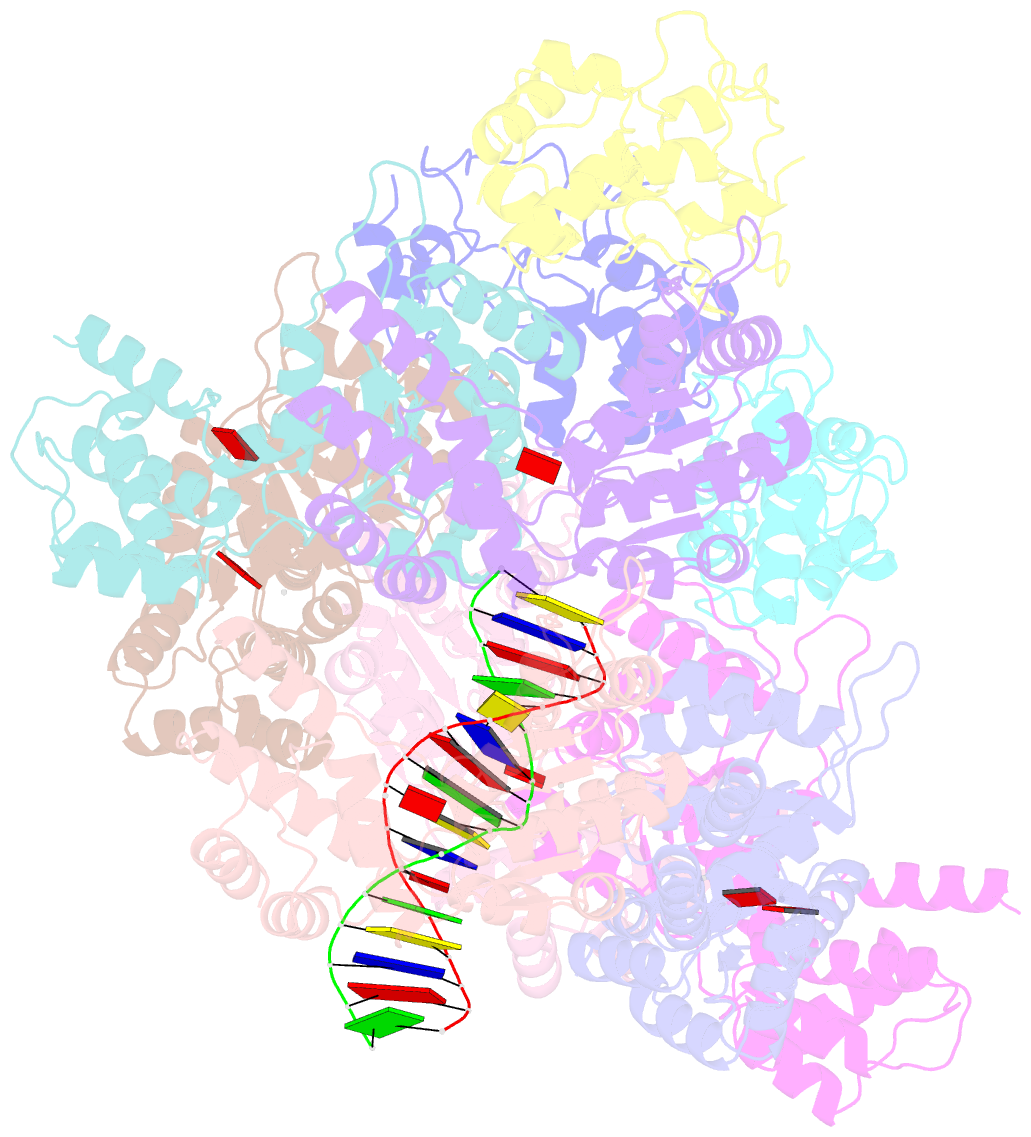
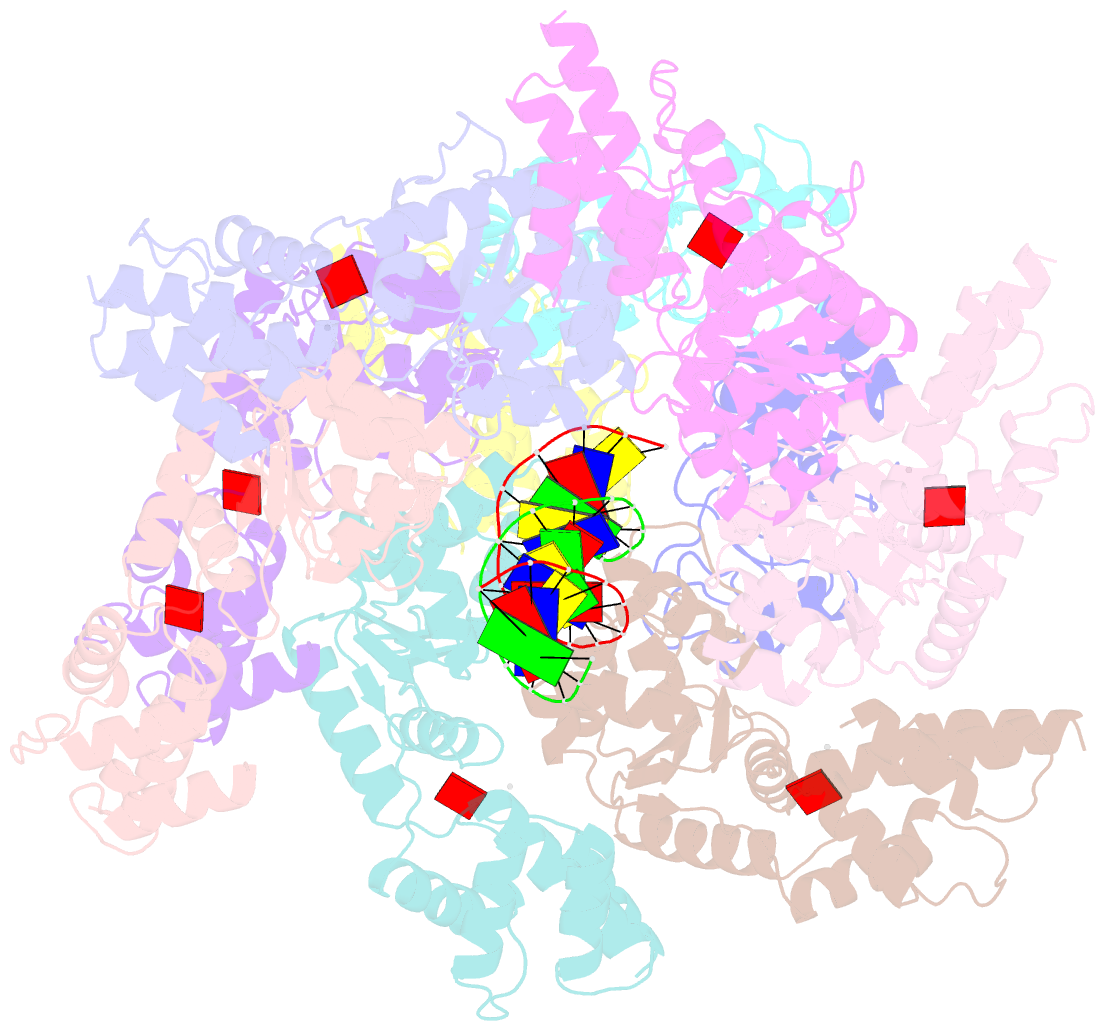
Summary information and primary citation
- PDB-id
- 8bd4; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- DNA binding protein
- Method
- cryo-EM (3.44 Å)
- Summary
- Tniq-capped tns-atp-dsDNA complex
- Reference
- Schmitz M, Querques I, Oberli S, Chanez C, Jinek M (2022): "Structural basis for the assembly of the type V CRISPR-associated transposon complex." Cell, 185, 4999. doi: 10.1016/j.cell.2022.11.009.
- Abstract
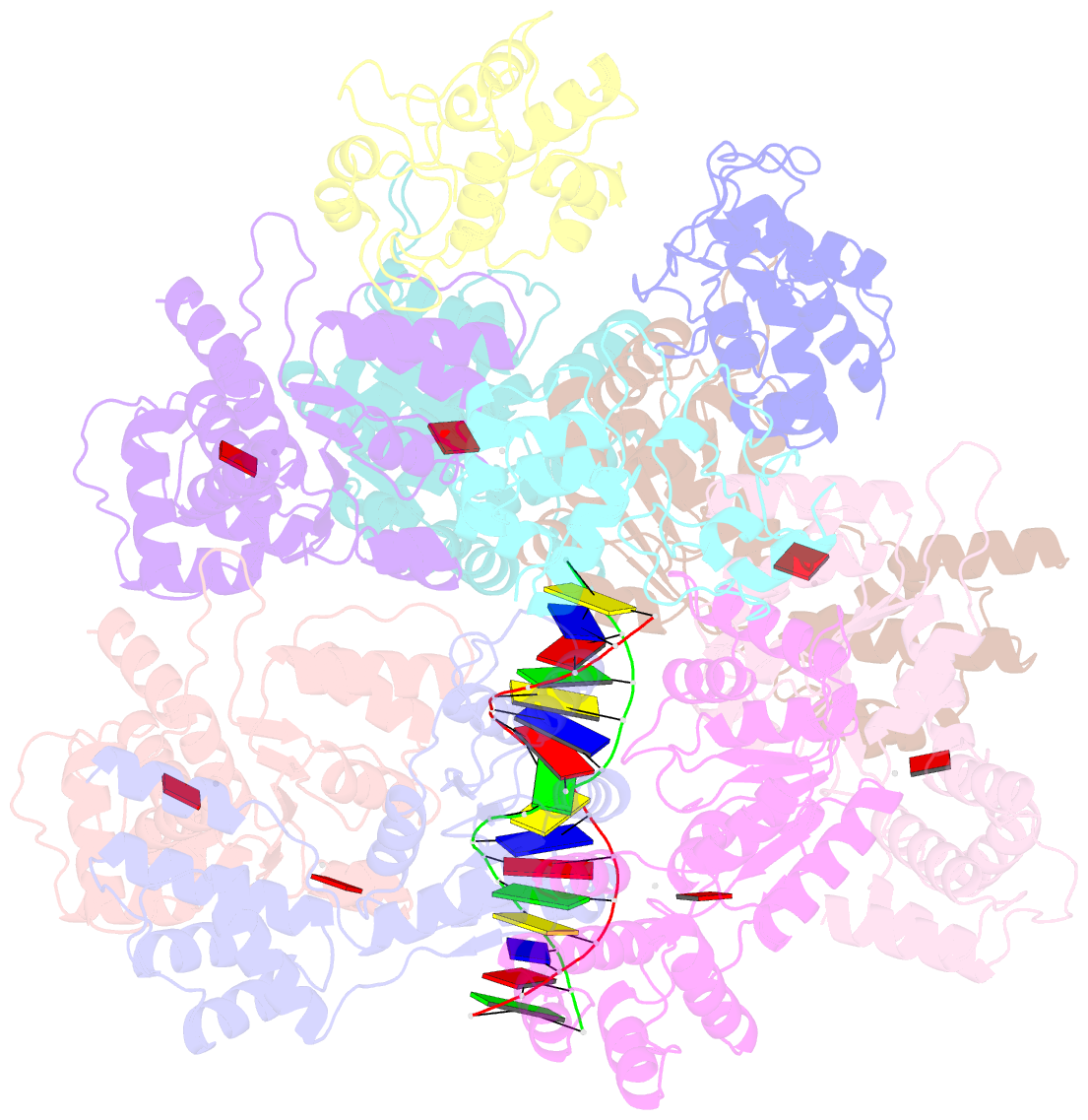
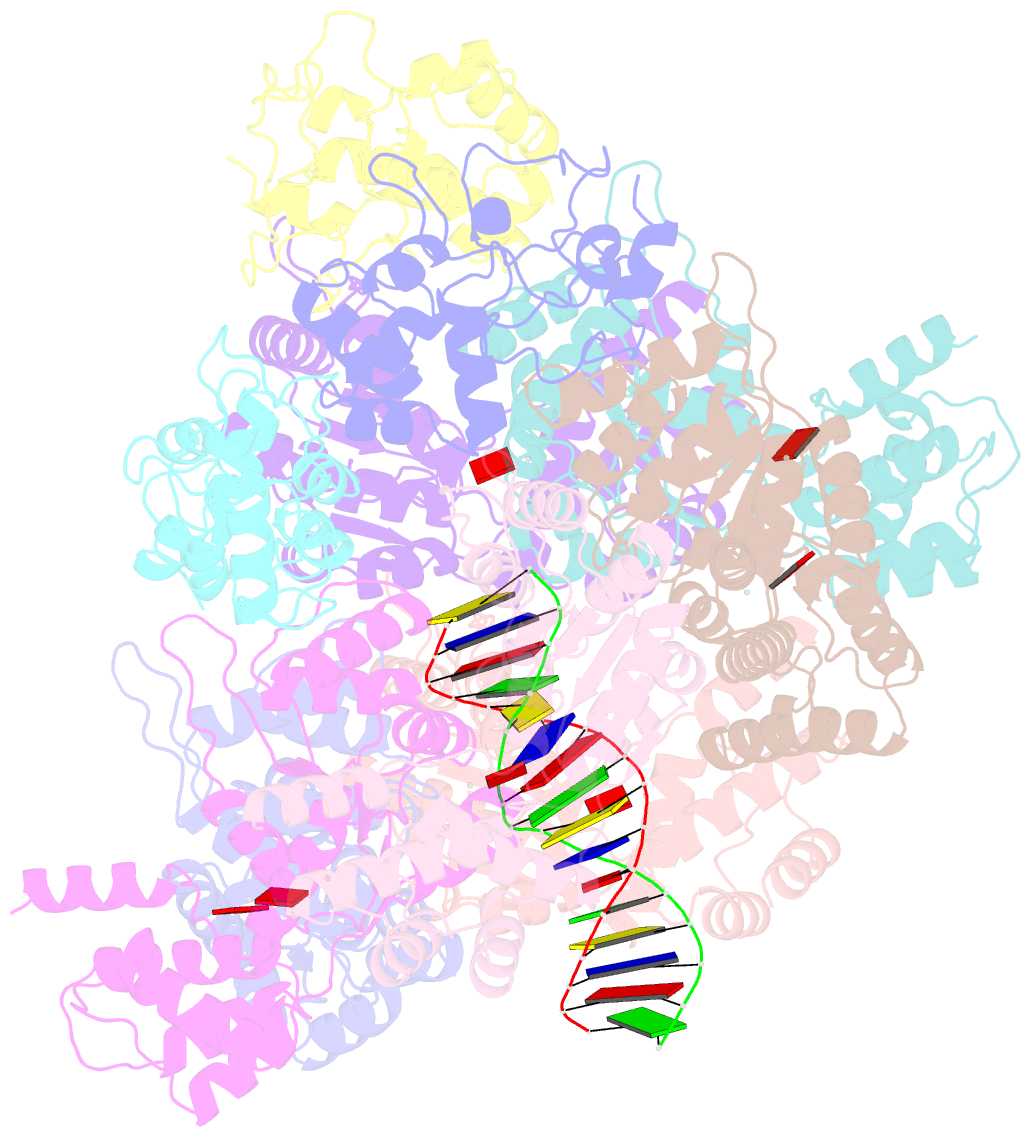
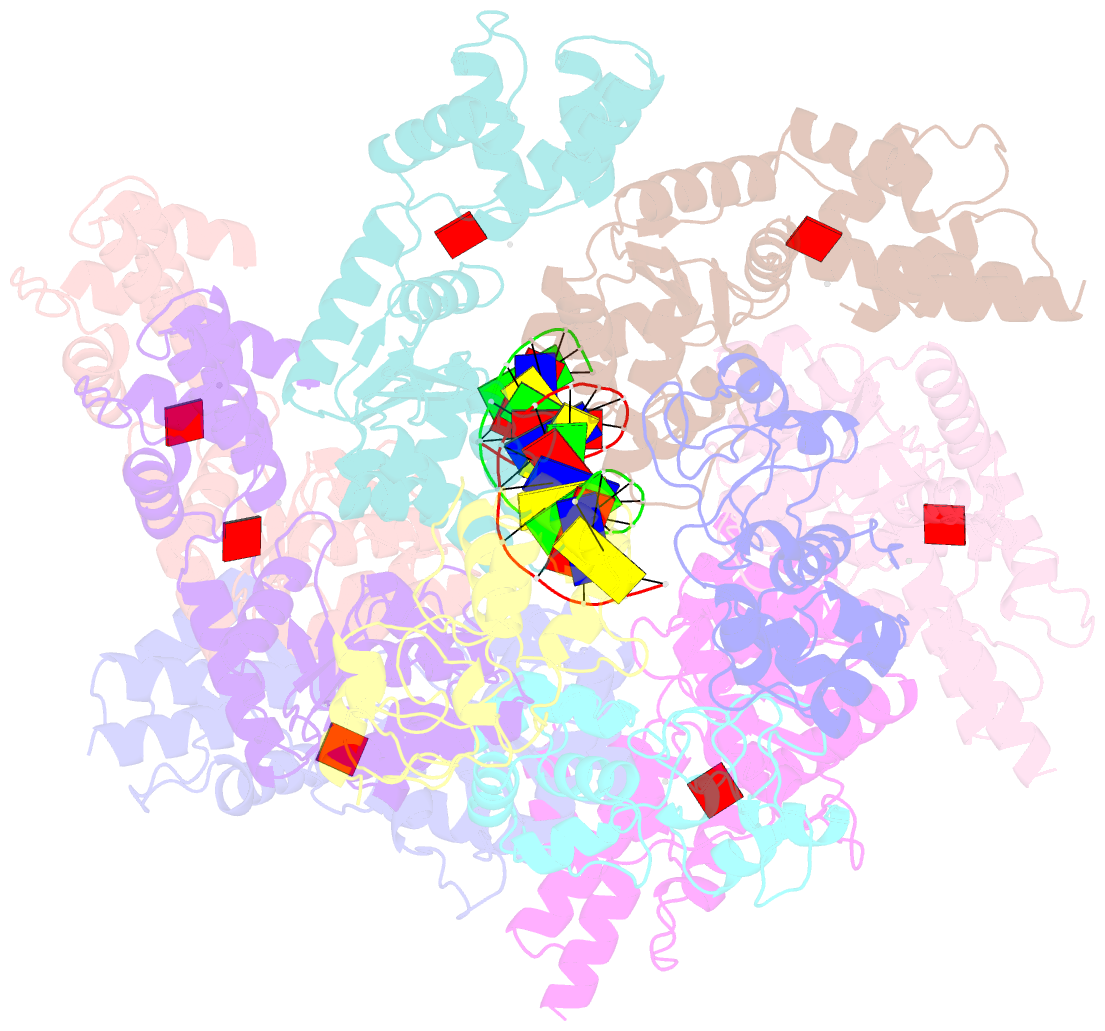
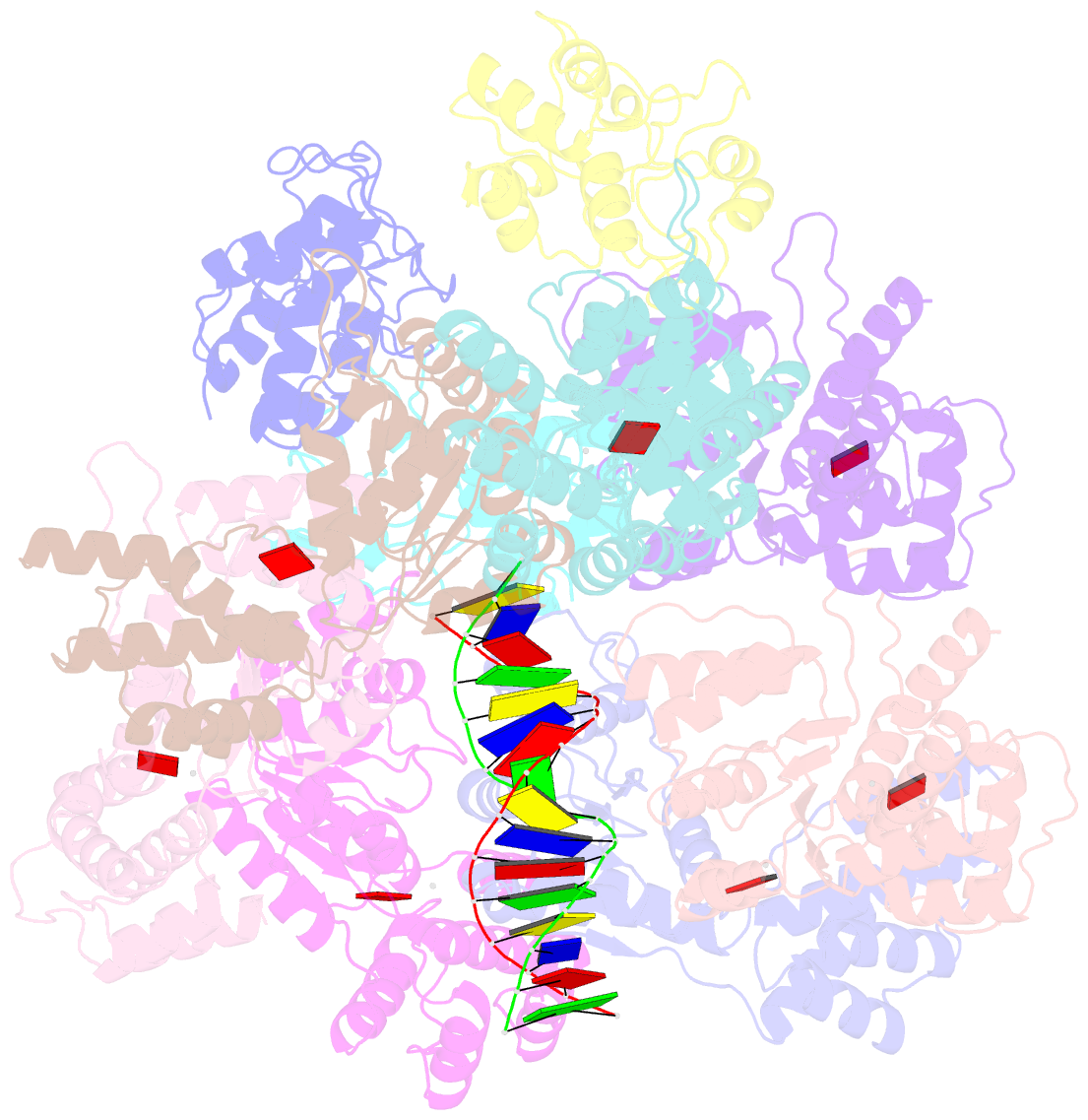
- CRISPR-Cas systems have been co-opted by Tn7-like transposable elements to direct RNA-guided transposition. Type V-K CRISPR-associated transposons rely on the concerted activities of the pseudonuclease Cas12k, the AAA+ ATPase TnsC, the Zn-finger protein TniQ, and the transposase TnsB. Here we present a cryo-electron microscopic structure of a target DNA-bound Cas12k-transposon recruitment complex comprised of RNA-guided Cas12k, TniQ, a polymeric TnsC filament and, unexpectedly, the ribosomal protein S15. Complex assembly, mediated by a network of interactions involving the guide RNA, TniQ, and S15, results in R-loop completion. TniQ contacts two TnsC protomers at the Cas12k-proximal filament end, likely nucleating its polymerization. Transposition activity assays corroborate our structural findings, implying that S15 is a bona fide component of the type V crRNA-guided transposon machinery. Altogether, our work uncovers key mechanistic aspects underpinning RNA-mediated assembly of CRISPR-associated transposons to guide their development as programmable tools for site-specific insertion of large DNA payloads.