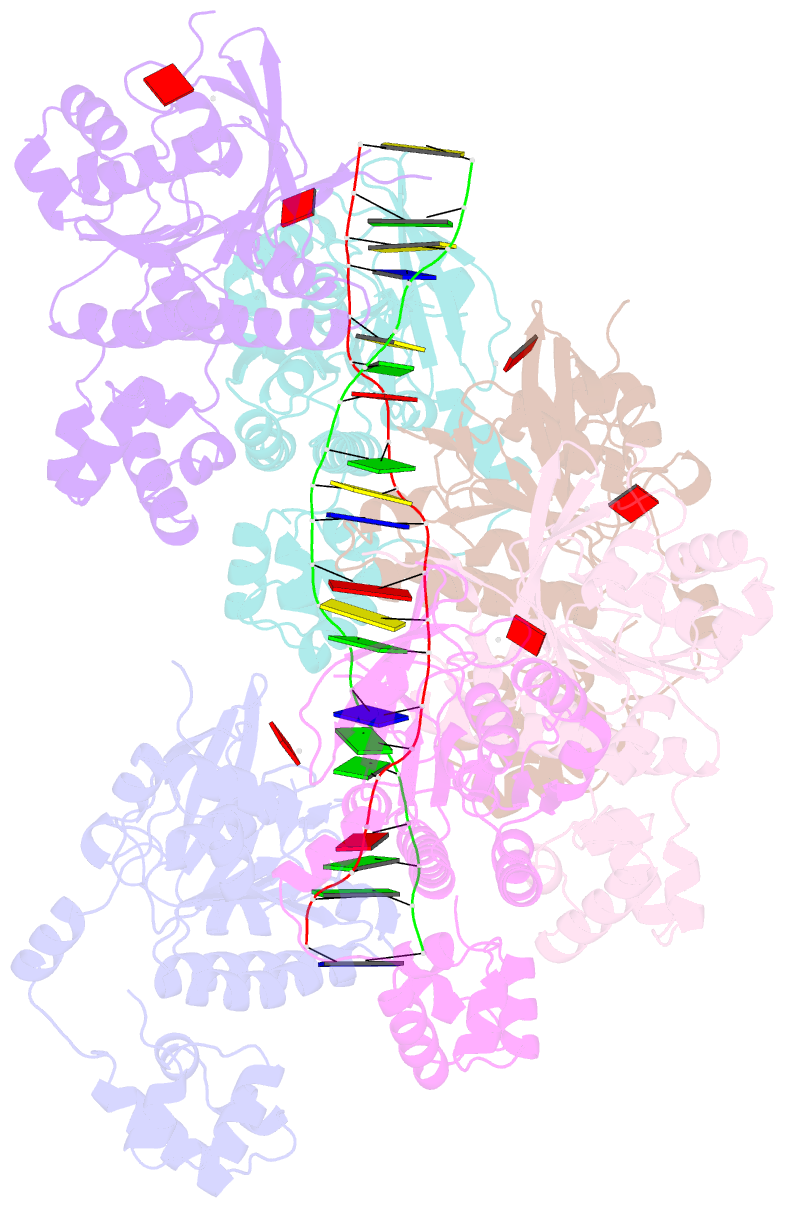
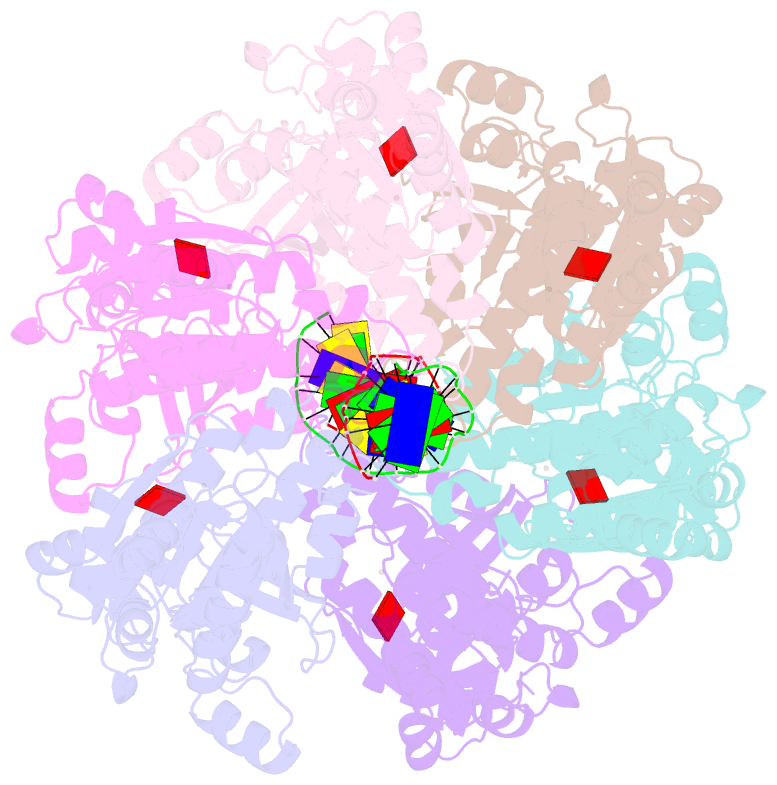
Summary information and primary citation
- PDB-id
- 8br2; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- DNA binding protein
- Method
- cryo-EM (2.9 Å)
- Summary
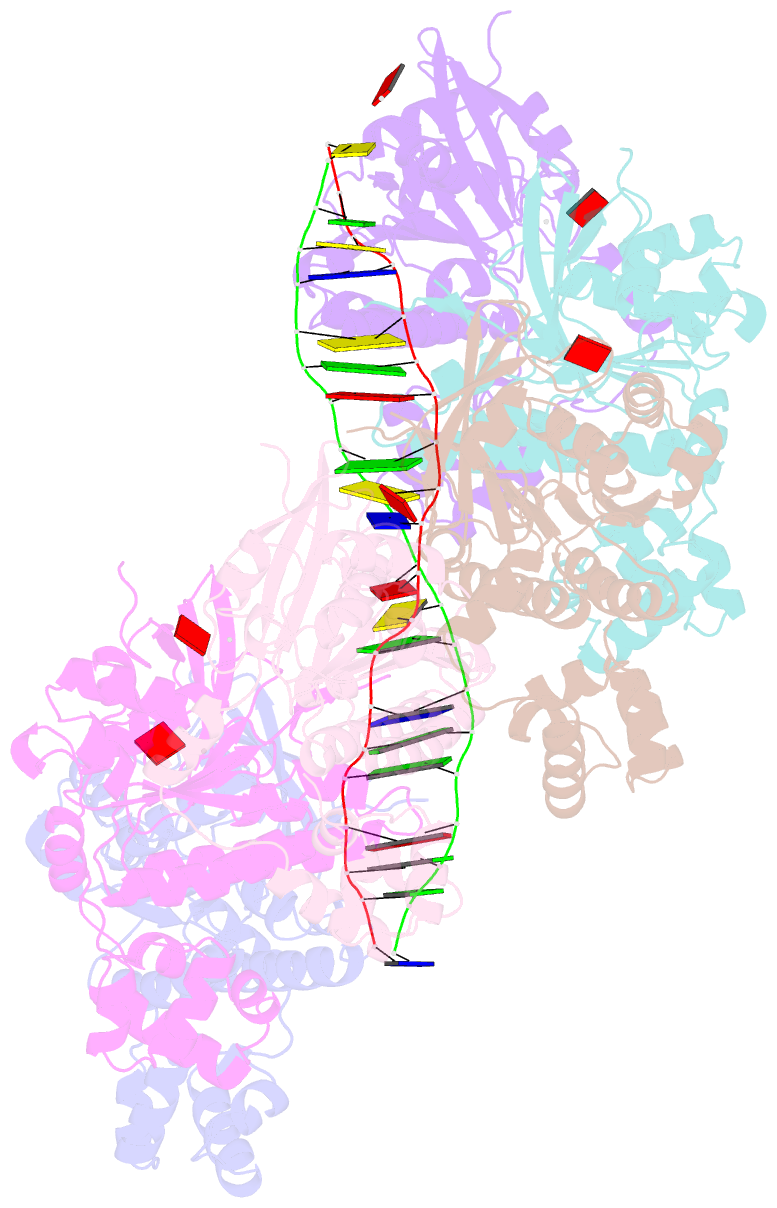
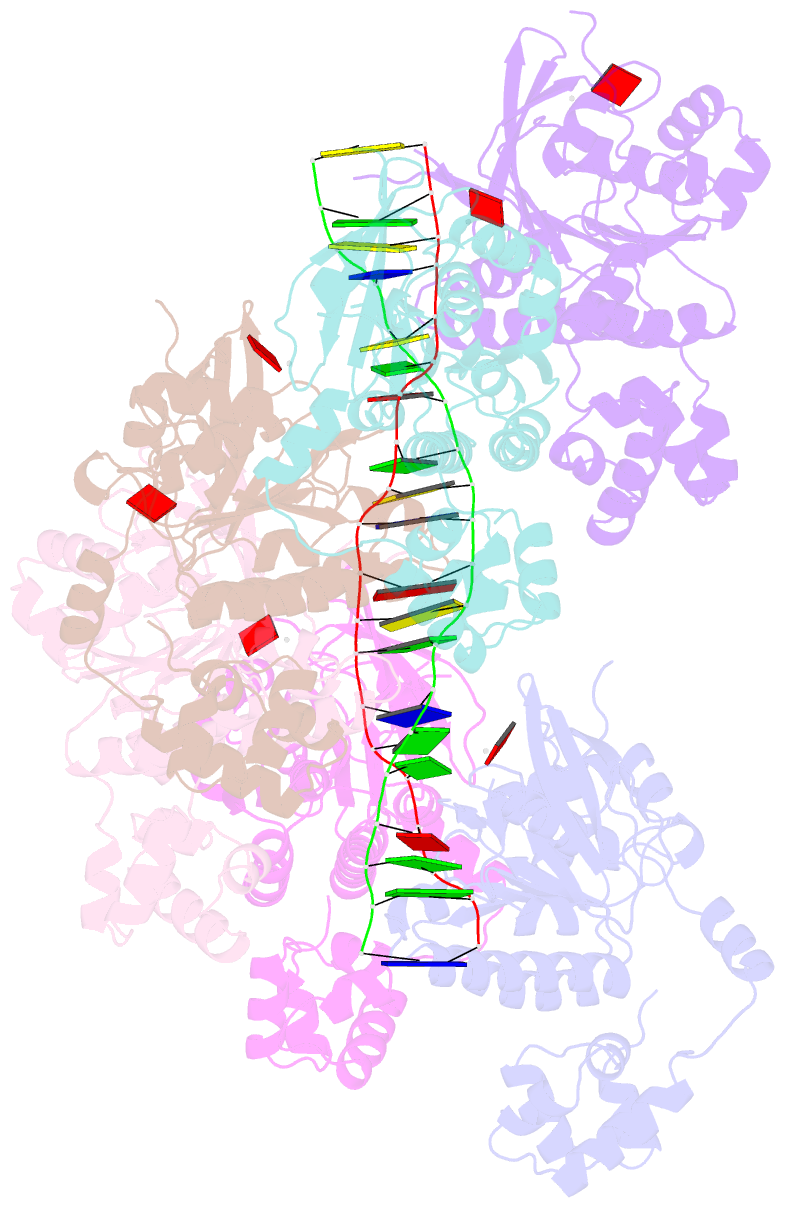
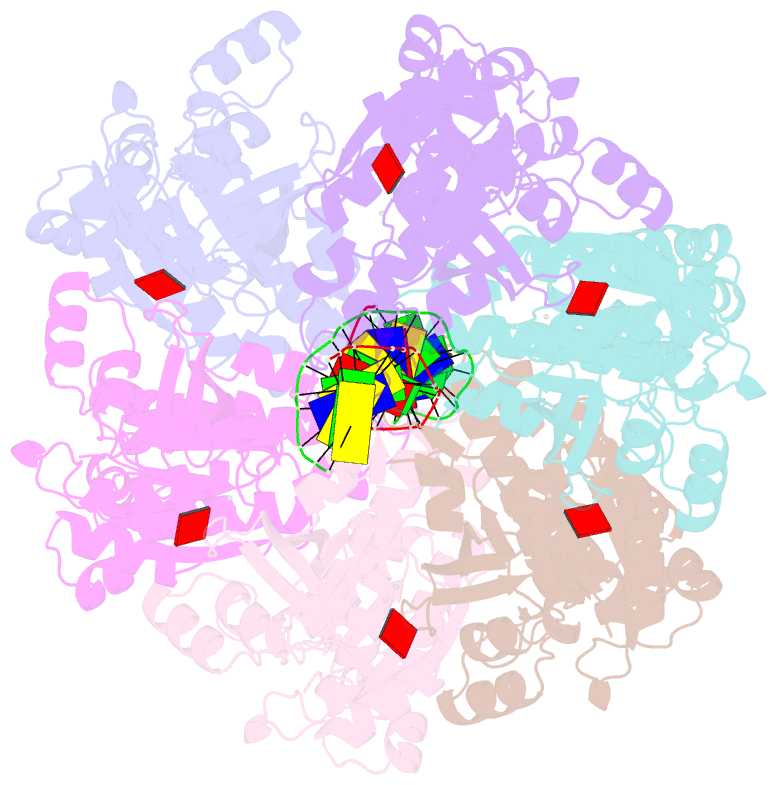
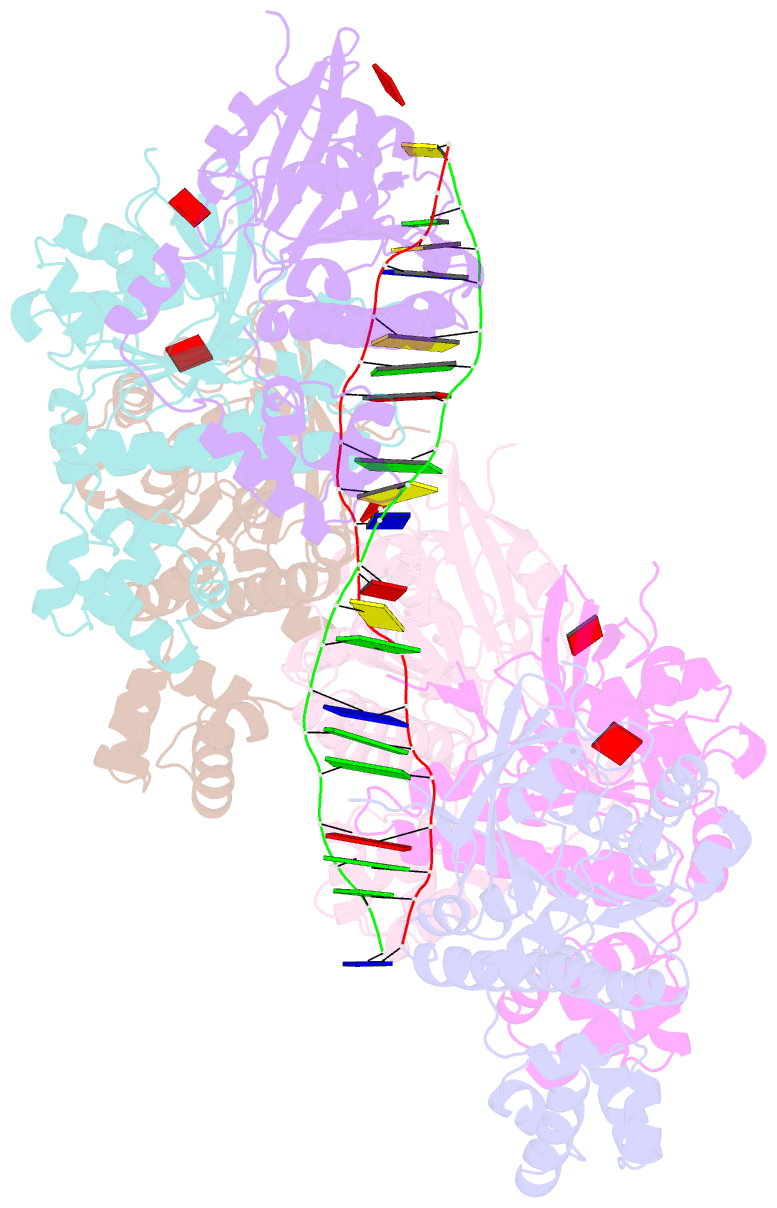
- Cryoem structure of the post-synaptic rad51 nucleoprotein filament in the presence of atp and ca2+
- Reference
- Appleby R, Bollschweiler D, Chirgadze DY, Joudeh L, Pellegrini L (2023): "A metal ion-dependent mechanism of RAD51 nucleoprotein filament disassembly." Iscience, 26, 106689. doi: 10.1016/j.isci.2023.106689.
- Abstract
- The RAD51 ATPase polymerizes on single-stranded DNA to form nucleoprotein filaments (NPFs) that are critical intermediates in the reaction of homologous recombination. ATP binding maintains the NPF in a competent conformation for strand pairing and exchange. Once strand exchange is completed, ATP hydrolysis licenses the filament for disassembly. Here we show that the ATP-binding site of the RAD51 NPF contains a second metal ion. In the presence of ATP, the metal ion promotes the local folding of RAD51 into the conformation required for DNA binding. The metal ion is absent in the ADP-bound RAD51 filament, that rearranges in a conformation incompatible with DNA binding. The presence of the second metal ion explains how RAD51 couples the nucleotide state of the filament to DNA binding. We propose that loss of the second metal ion upon ATP hydrolysis drives RAD51 dissociation from the DNA and weakens filament stability, contributing to NPF disassembly.