Summary information and primary citation
- PDB-id
- 8btg; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- replication
- Method
- cryo-EM (3.2 Å)
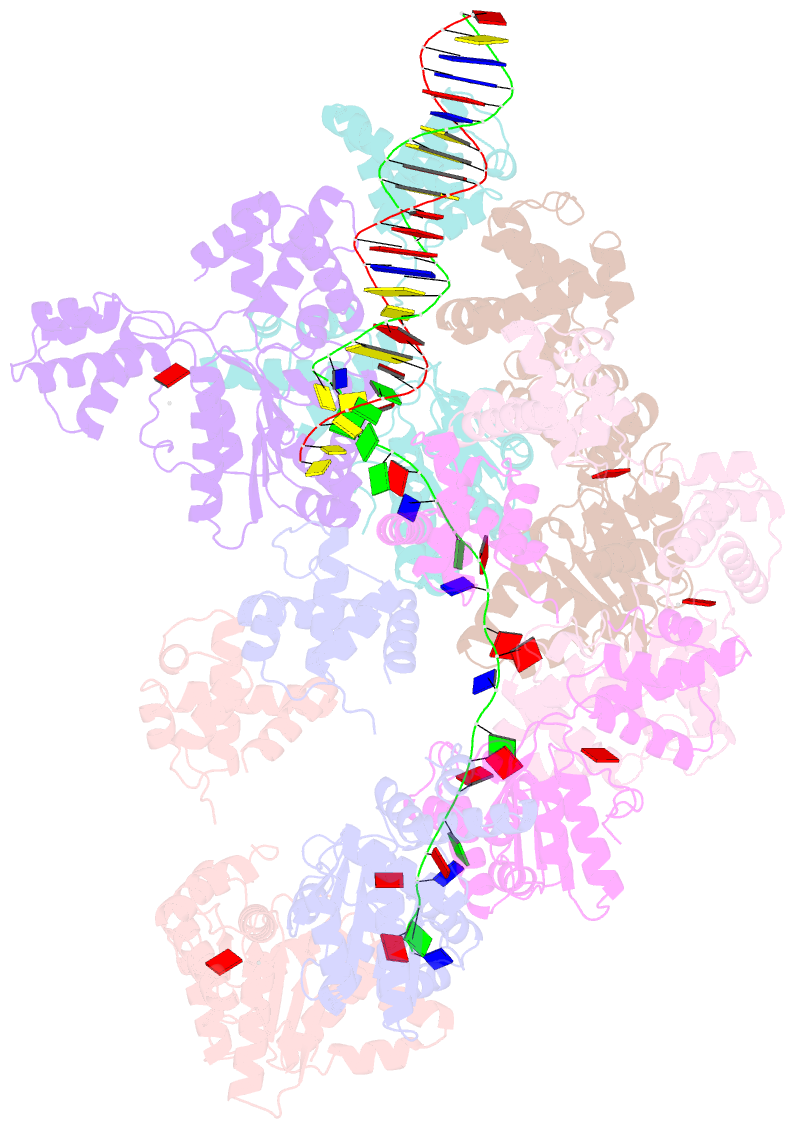
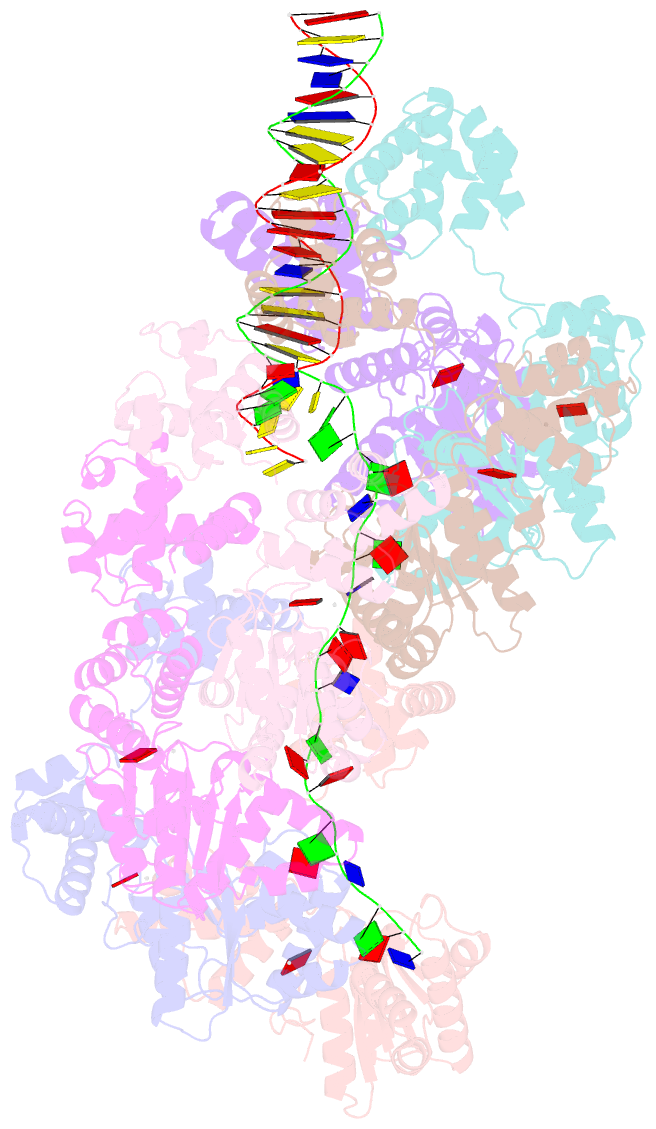
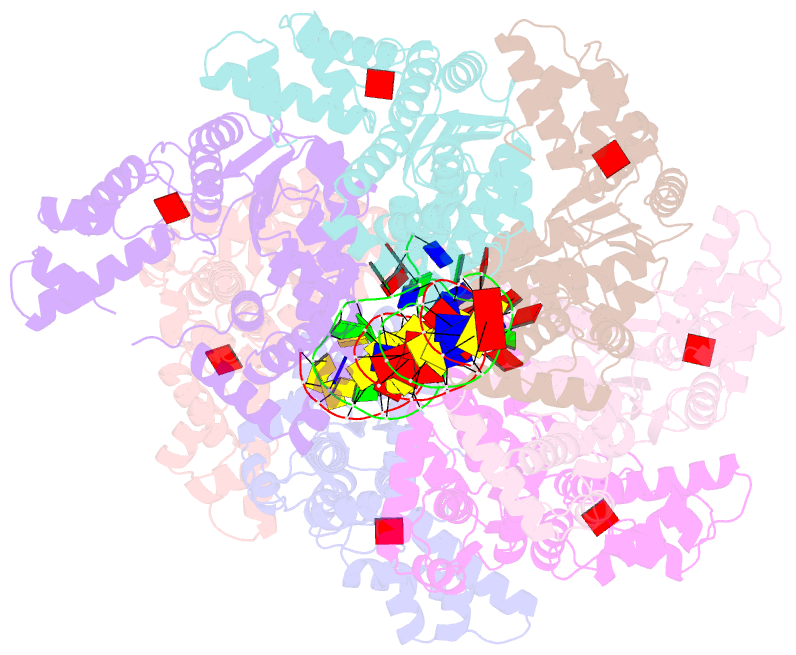
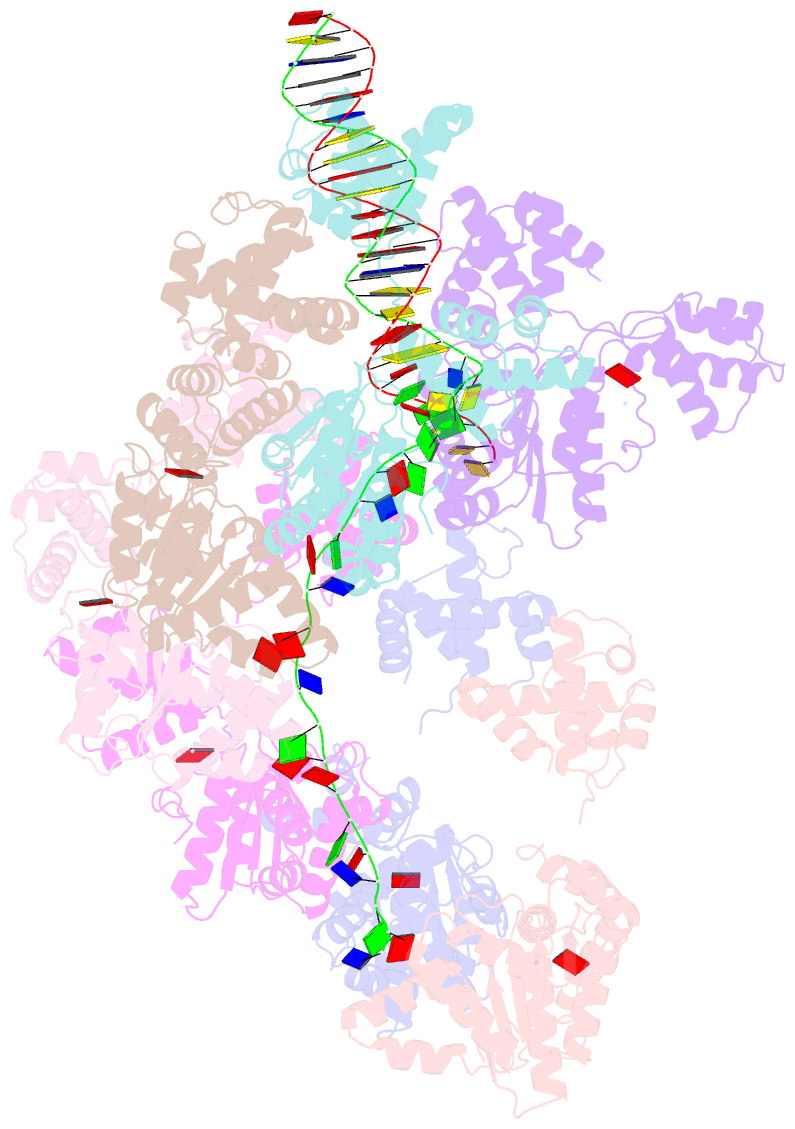
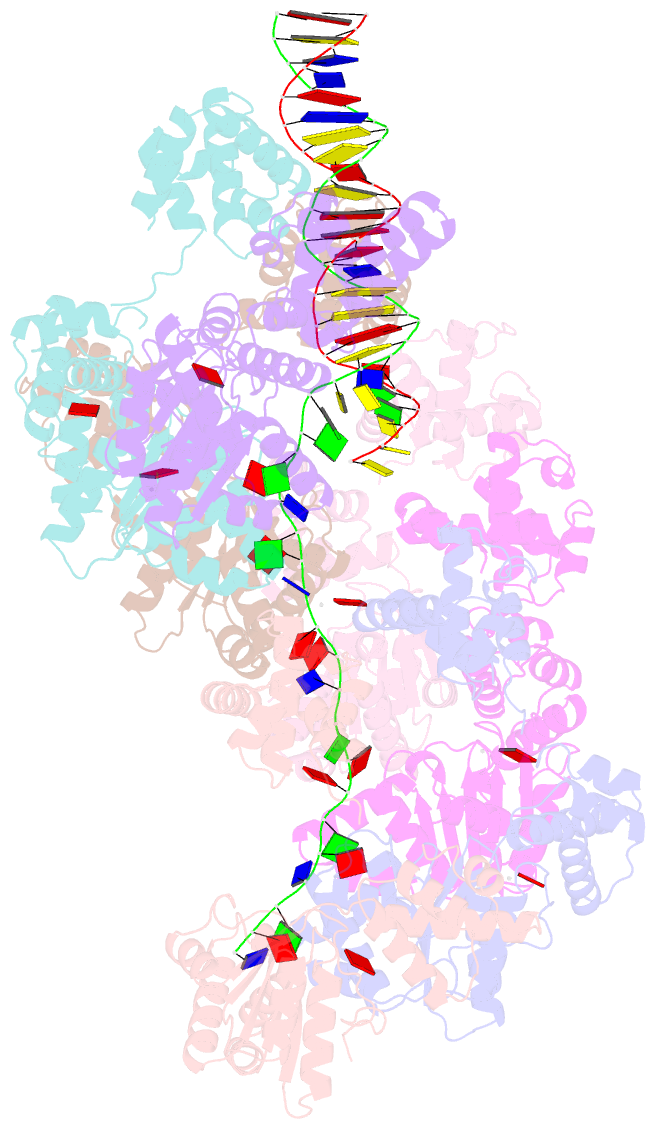
- Summary
- cryo-EM structure of the bacterial replication origin opening basal unwinding system
- Reference
- Pelliciari S, Bodet-Lefevre S, Fenyk S, Stevens D, Winterhalter C, Schramm FD, Pintar S, Burnham DR, Merces G, Richardson TT, Tashiro Y, Hubbard J, Yardimci H, Ilangovan A, Murray H (2023): "The bacterial replication origin BUS promotes nucleobase capture." Nat Commun, 14, 8339. doi: 10.1093/nar/gkac1060.
- Abstract
- Genome duplication is essential for the proliferation of cellular life and this process is generally initiated by dedicated replication proteins at chromosome origins. In bacteria, DNA replication is initiated by the ubiquitous DnaA protein, which assembles into an oligomeric complex at the chromosome origin (oriC) that engages both double-stranded DNA (dsDNA) and single-stranded DNA (ssDNA) to promote DNA duplex opening. However, the mechanism of DnaA specifically opening a replication origin was unknown. Here we show that Bacillus subtilis DnaAATP assembles into a continuous oligomer at the site of DNA melting, extending from a dsDNA anchor to engage a single DNA strand. Within this complex, two nucleobases of each ssDNA binding motif (DnaA-trio) are captured within a dinucleotide binding pocket created by adjacent DnaA proteins. These results provide a molecular basis for DnaA specifically engaging the conserved sequence elements within the bacterial chromosome origin basal unwinding system (BUS).