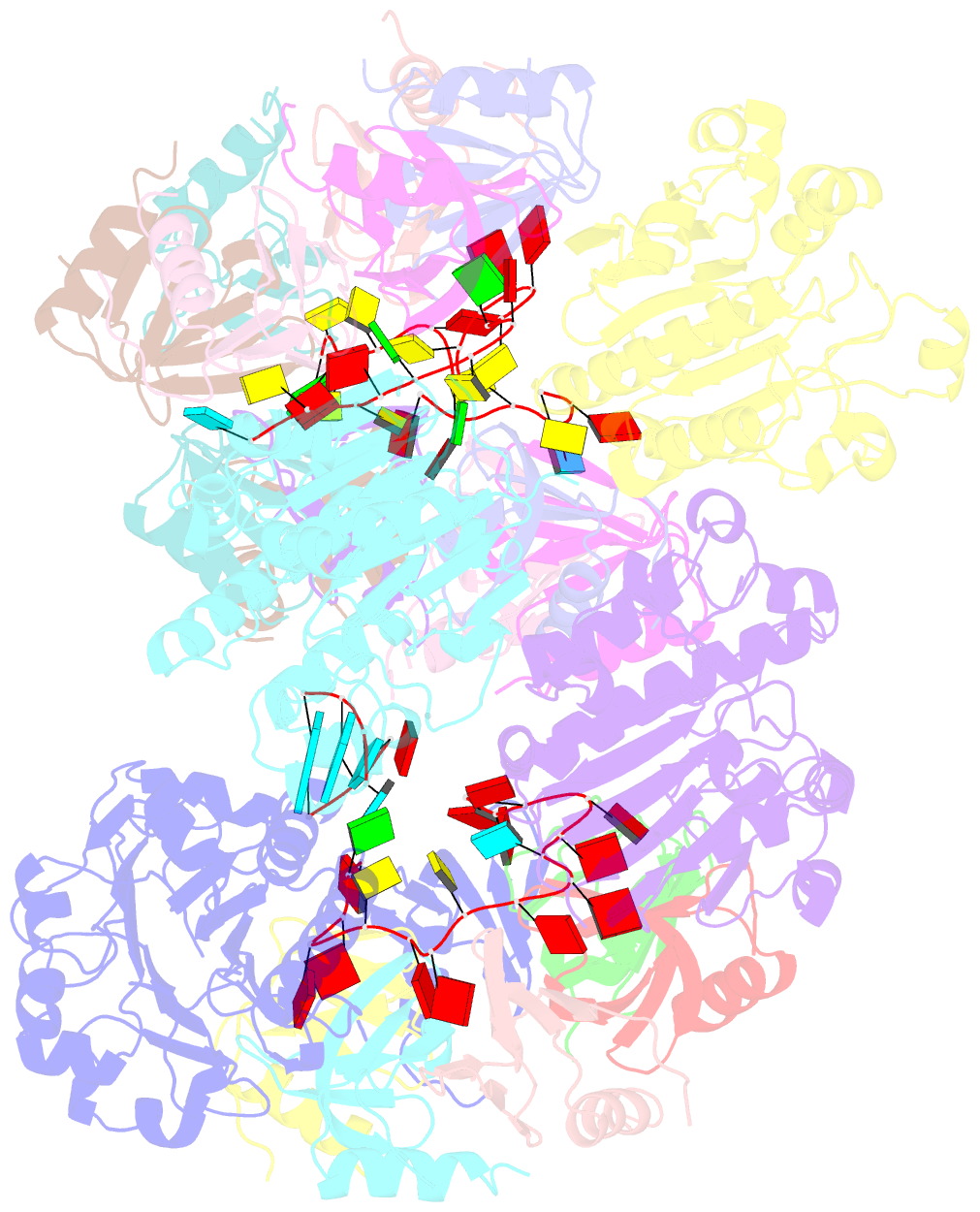
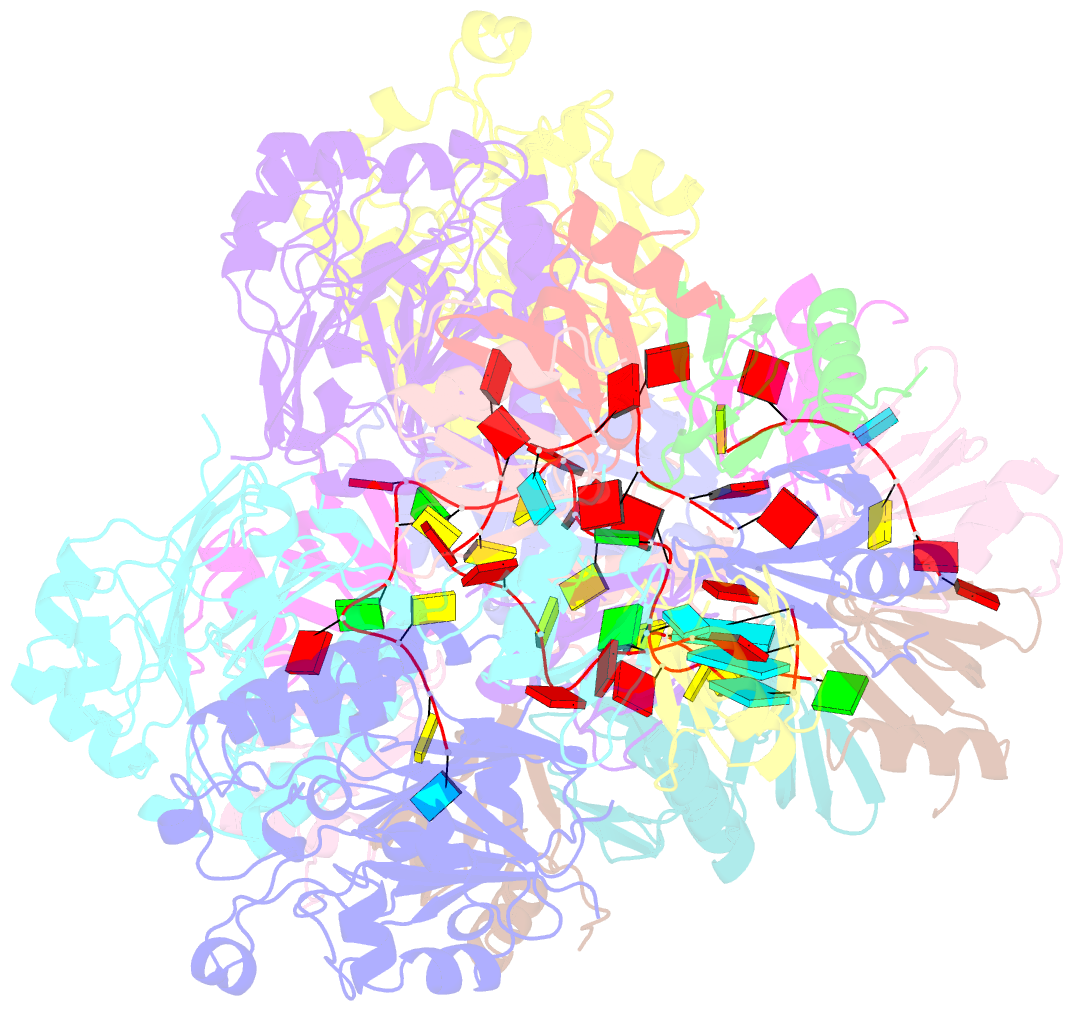
Summary information and primary citation
- PDB-id
- 8bvh; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- RNA binding protein
- Method
- cryo-EM (3.6 Å)
- Summary
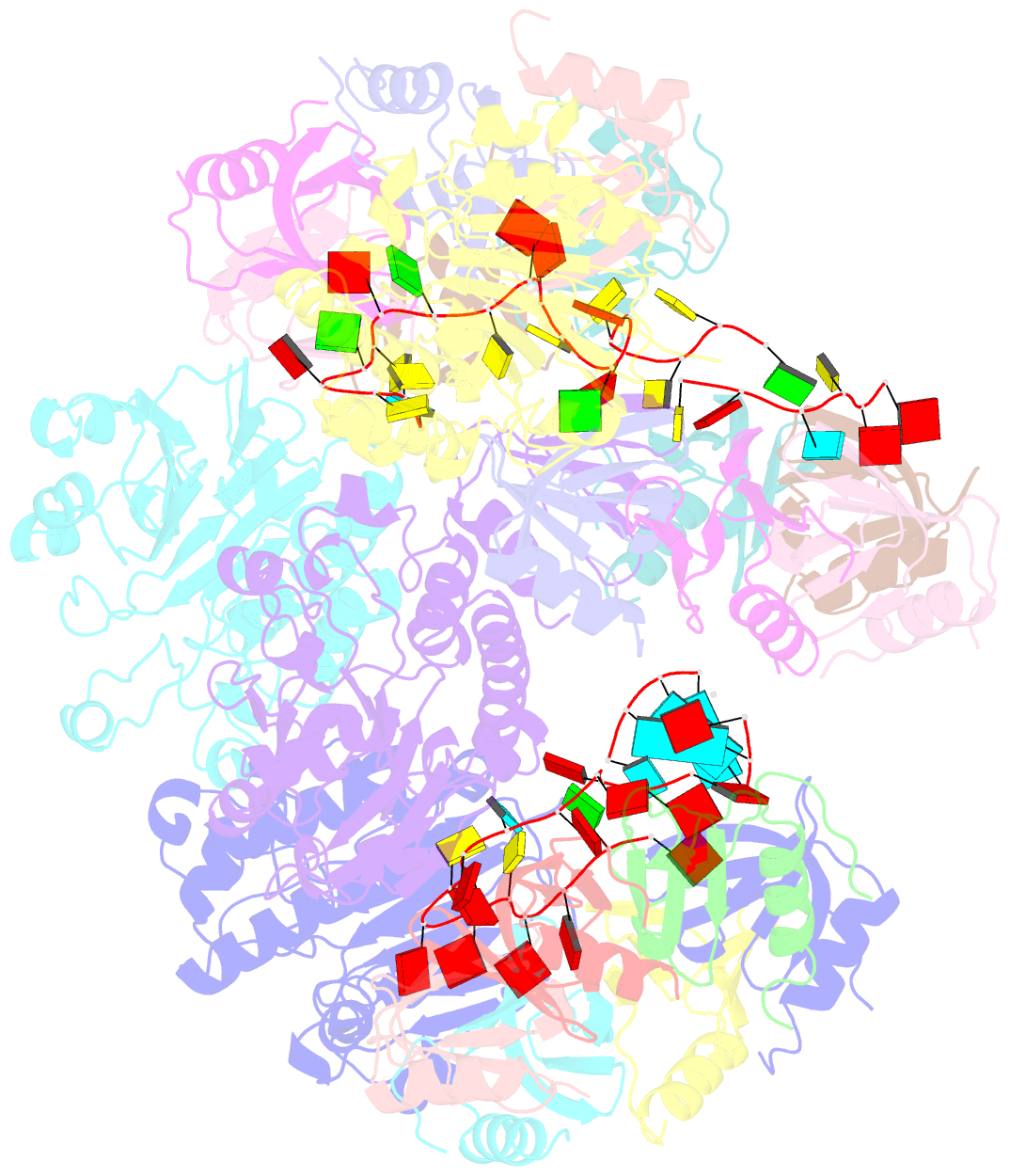
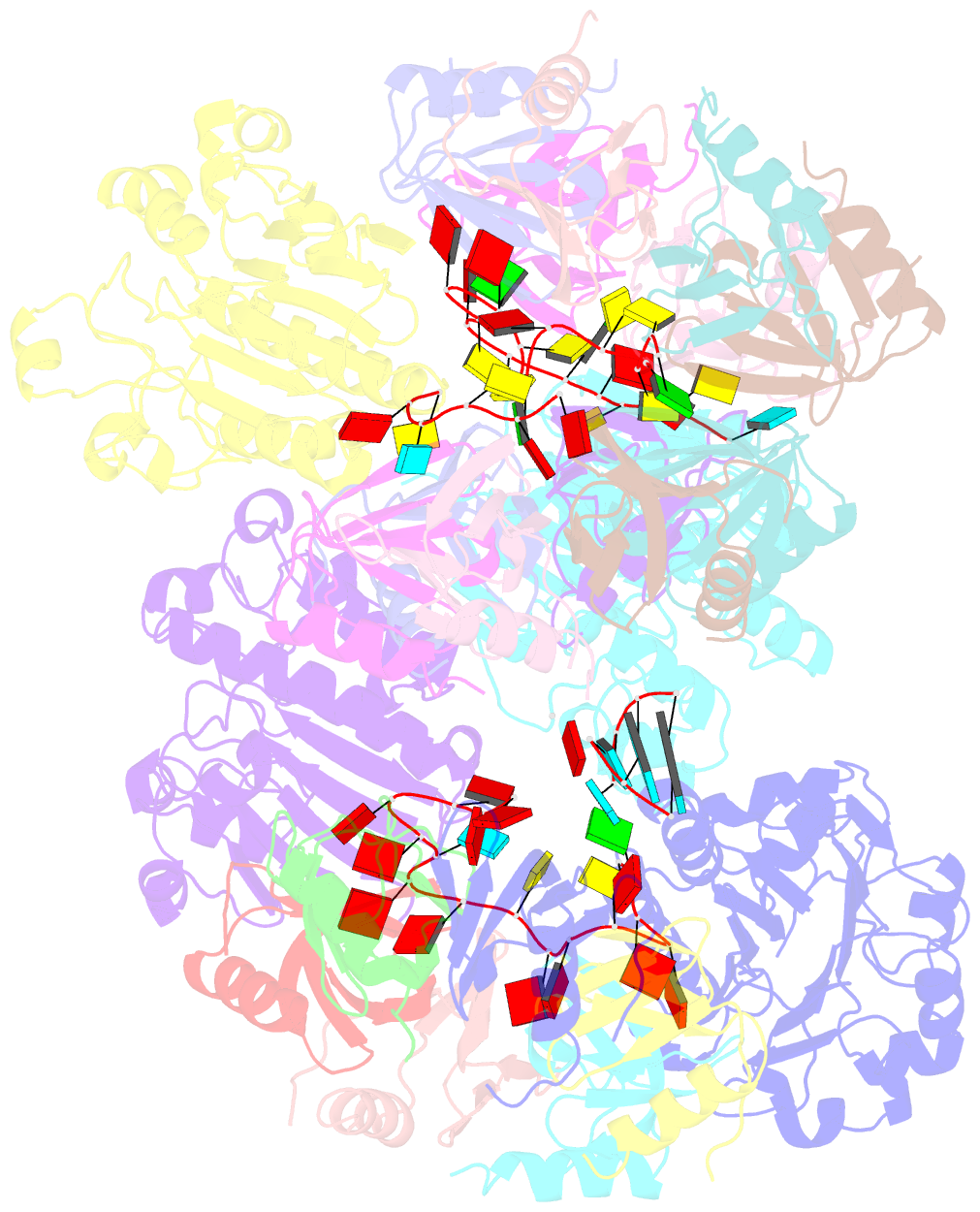
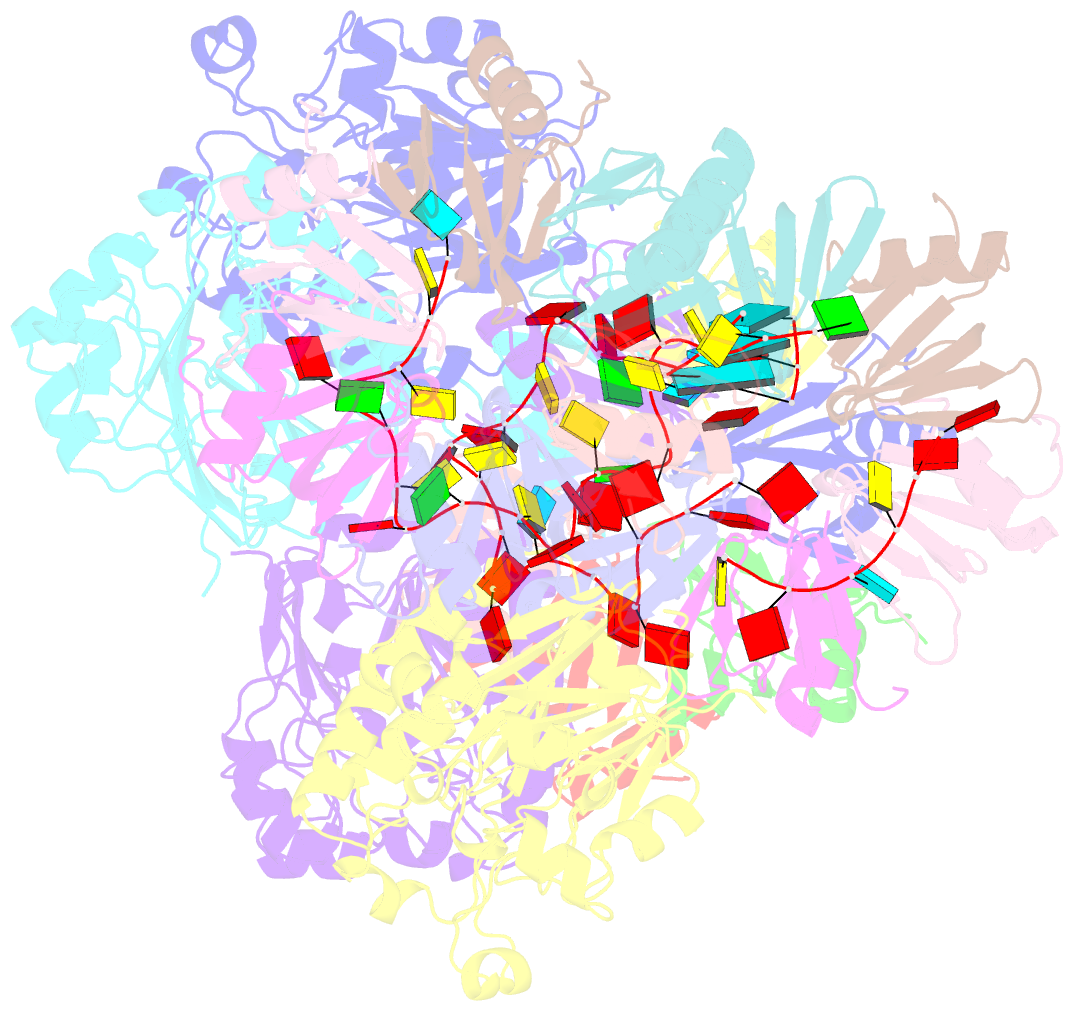
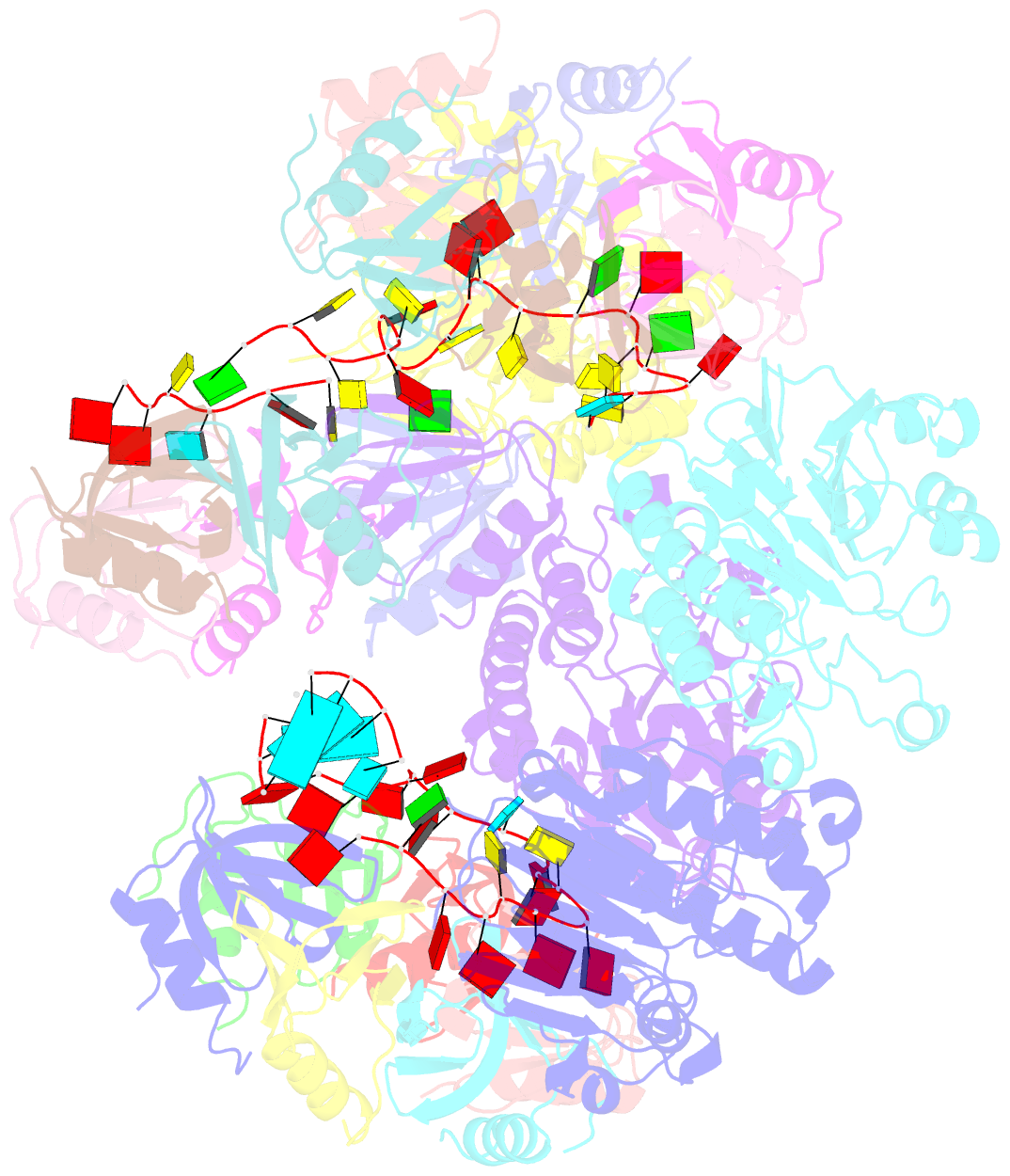
- cryo-EM structure of the hfq-crc-amie translation repression assembly.
- Reference
- Dendooven T, Sonnleitner E, Blasi U, Luisi BF (2023): "Translational regulation by Hfq-Crc assemblies emerges from polymorphic ribonucleoprotein folding." Embo J., 42, e111129. doi: 10.15252/embj.2022111129.
- Abstract
- The widely occurring bacterial RNA chaperone Hfq is a key factor in the post-transcriptional control of hundreds of genes in Pseudomonas aeruginosa. How this broadly acting protein can contribute to the regulatory requirements of many different genes remains puzzling. Here, we describe cryo-EM structures of higher order assemblies formed by Hfq and its partner protein Crc on control regions of different P. aeruginosa target mRNAs. Our results show that these assemblies have mRNA-specific quaternary architectures resulting from the combination of multivalent protein-protein interfaces and recognition of patterns in the RNA sequence. The structural polymorphism of these ribonucleoprotein assemblies enables selective translational repression of many different target mRNAs. This system elucidates how highly complex regulatory pathways can evolve with a minimal economy of proteinogenic components in combination with RNA sequence and fold.