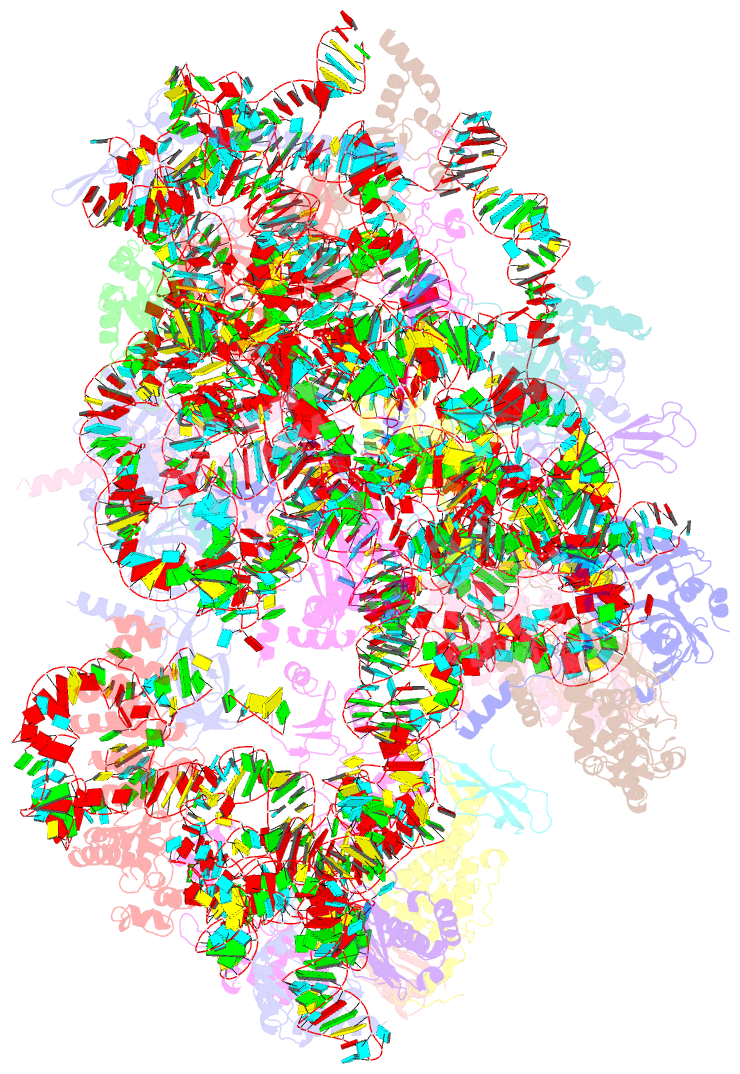
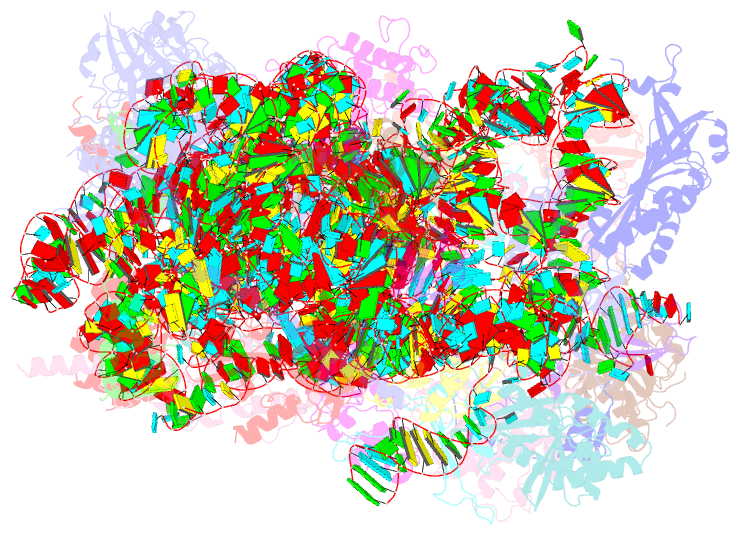
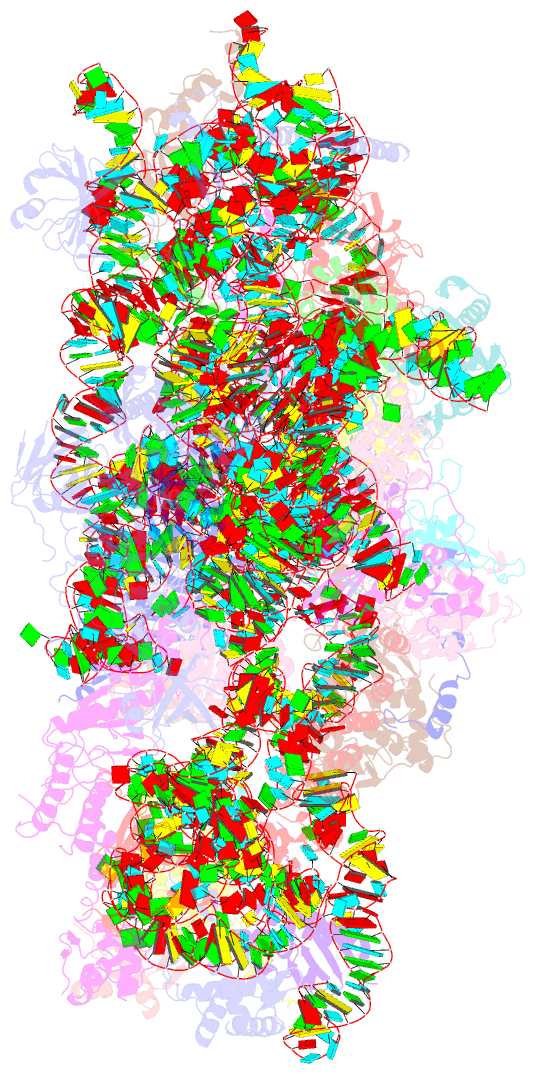
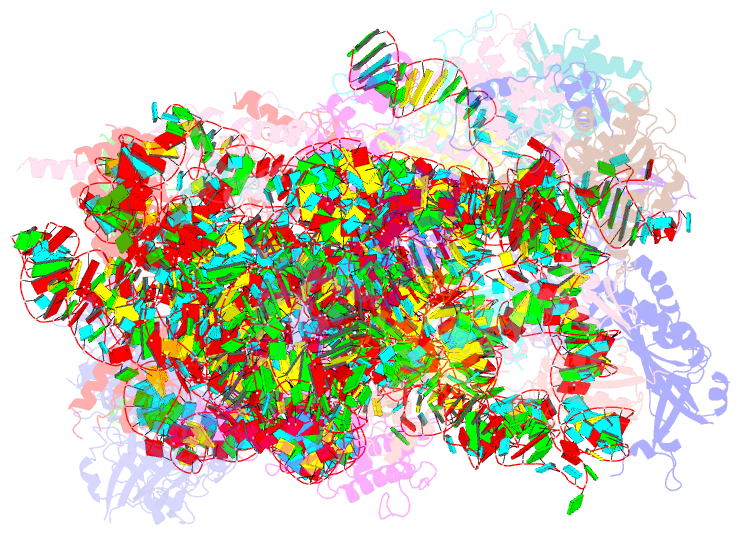
Summary information and primary citation
- PDB-id
- 8c01; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- ribosome
- Method
- cryo-EM (2.7 Å)
- Summary
- Enp1tap_a population of yeast small ribosomal subunit precursors
- Reference
- Poll G, Griesenbeck J, Tschochner H, Milkereit P (2023): "Impact of the yeast S0/uS2-cluster ribosomal protein rpS21/eS21 on rRNA folding and the architecture of small ribosomal subunit precursors." Plos One, 18, e0283698. doi: 10.1371/journal.pone.0283698.
- Abstract
- RpS0/uS2, rpS2/uS5, and rpS21/eS21 form a cluster of ribosomal proteins (S0-cluster) at the head-body junction near the central pseudoknot of eukaryotic small ribosomal subunits (SSU). Previous work in yeast indicated that S0-cluster assembly is required for the stabilisation and maturation of SSU precursors at specific post-nucleolar stages. Here, we analysed the role of S0-cluster formation for rRNA folding. Structures of SSU precursors isolated from yeast S0-cluster expression mutants or control strains were analysed by cryogenic electron microscopy. The obtained resolution was sufficient to detect individual 2'-O-methyl RNA modifications using an unbiased scoring approach. The data show how S0-cluster formation enables the initial recruitment of the pre-rRNA processing factor Nob1 in yeast. Furthermore, they reveal hierarchical effects on the pre-rRNA folding pathway, including the final maturation of the central pseudoknot. Based on these structural insights we discuss how formation of the S0-cluster determines at this early cytoplasmic assembly checkpoint if SSU precursors further mature or are degraded.