Summary information and primary citation
- PDB-id
- 8cbw; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- viral protein
- Method
- cryo-EM (3.485 Å)
- Summary
- Cryoem structure of the hendra henipavirus nucleocapsid sauronoid assembly monomer
- Reference
- Passchier TC, White JBR, Maskell DP, Byrne MJ, Ranson NA, Edwards TA, Barr JN (2024): "The cryoEM structure of the Hendra henipavirus nucleoprotein reveals insights into paramyxoviral nucleocapsid architectures." Sci Rep, 14, 14099. doi: 10.1038/s41598-024-58243-z.
- Abstract
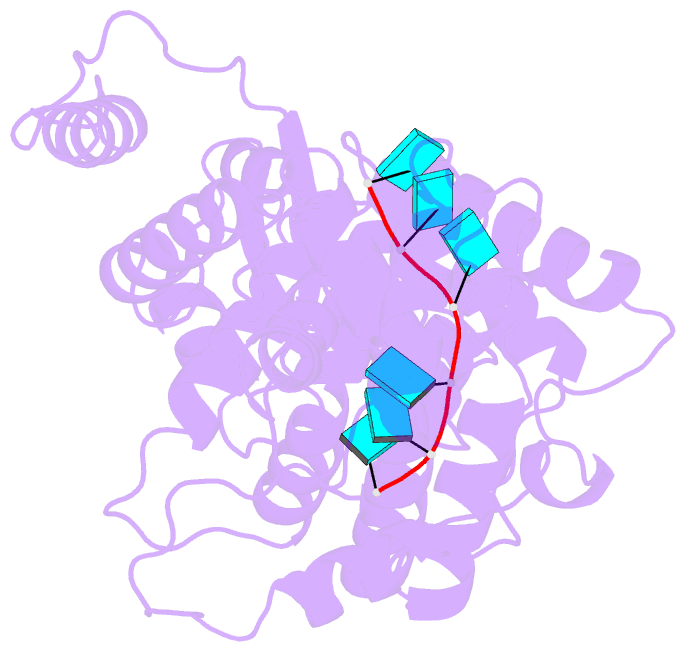
- We report the first cryoEM structure of the Hendra henipavirus nucleoprotein in complex with RNA, at 3.5 Å resolution, derived from single particle analysis of a double homotetradecameric RNA-bound N protein ring assembly exhibiting D14 symmetry. The structure of the HeV N protein adopts the common bi-lobed paramyxoviral N protein fold; the N-terminal and C-terminal globular domains are bisected by an RNA binding cleft containing six RNA nucleotides and are flanked by the N-terminal and C-terminal arms, respectively. In common with other paramyxoviral nucleocapsids, the lateral interface between adjacent Ni and Ni+1 protomers involves electrostatic and hydrophobic interactions mediated primarily through the N-terminal arm and globular domains with minor contribution from the C-terminal arm. However, the HeV N multimeric assembly uniquely identifies an additional protomer-protomer contact between the Ni+1 N-terminus and Ni-1 C-terminal arm linker. The model presented here broadens the understanding of RNA-bound paramyxoviral nucleocapsid architectures and provides a platform for further insight into the molecular biology of HeV, as well as the development of antiviral interventions.