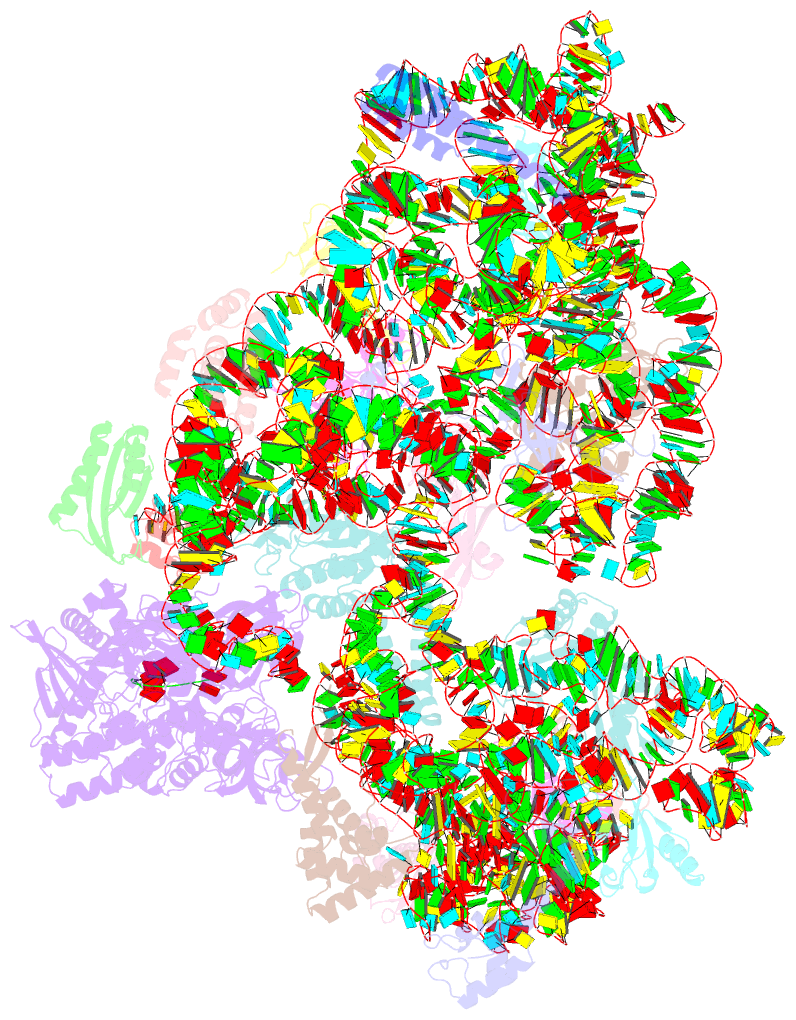
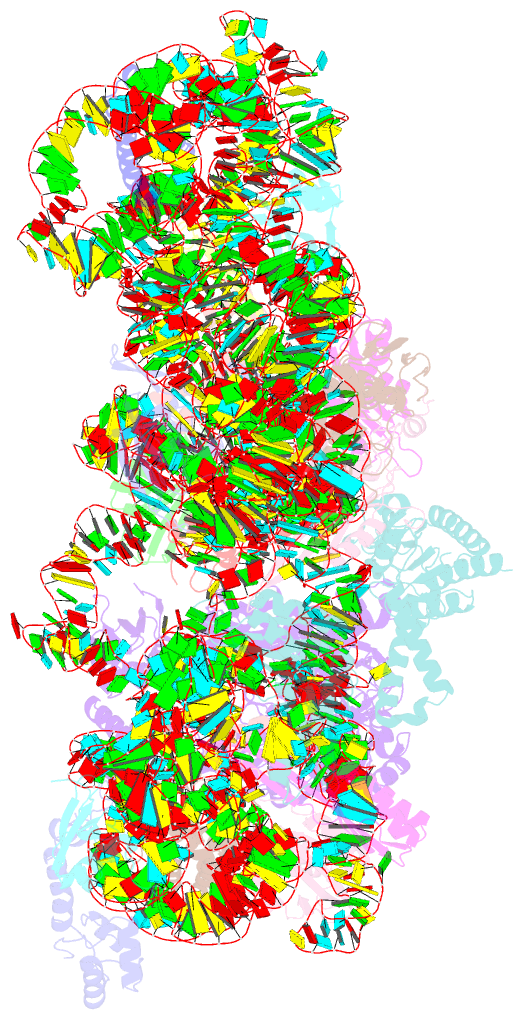
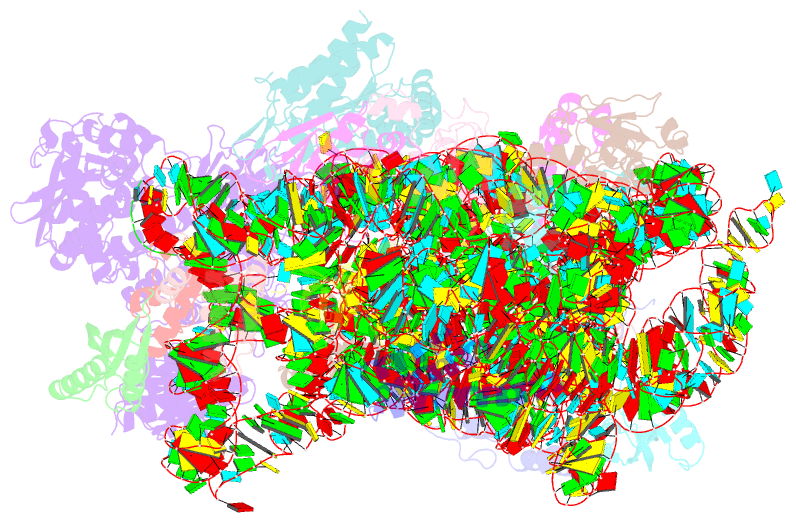
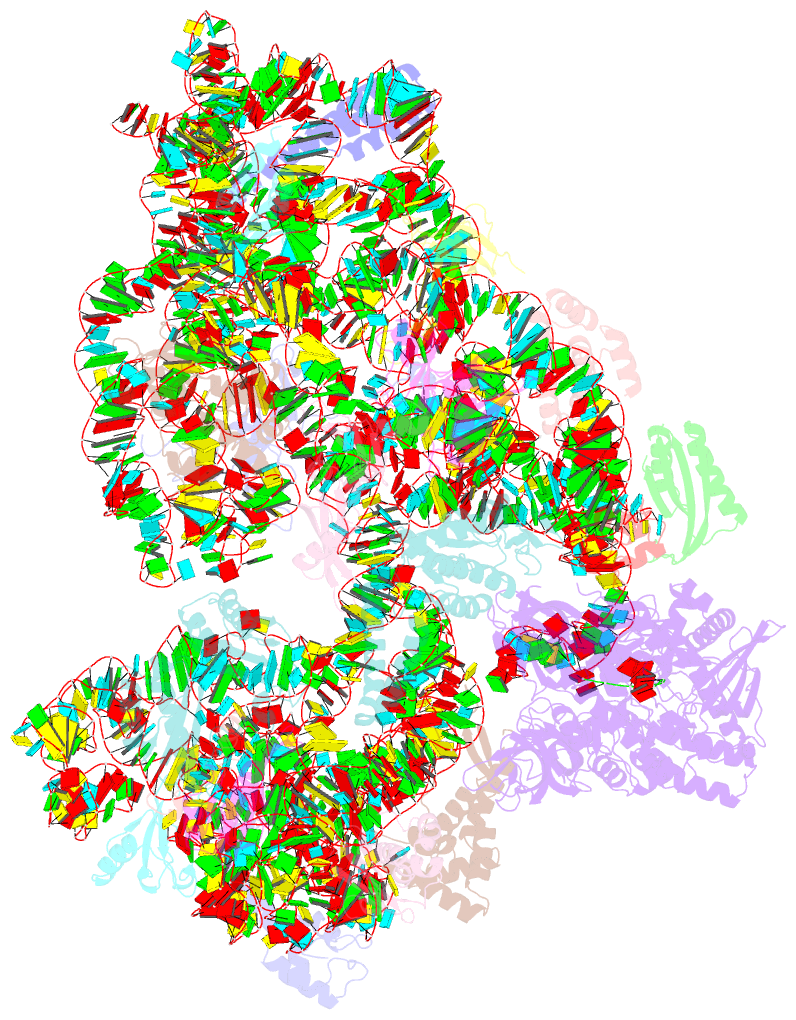
Summary information and primary citation
- PDB-id
- 8ced; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- ribosome
- Method
- cryo-EM (4.15 Å)
- Summary
- Rnase r bound to a 30s degradation intermediate (state i - head-turning)
- Reference
- Dimitrova-Paternoga L, Kasvandik S, Beckert B, Granneman S, Tenson T, Wilson DN, Paternoga H (2024): "Structural basis of ribosomal 30S subunit degradation by RNase R." Nature, 626, 1133-1140. doi: 10.1038/s41586-024-07027-6.
- Abstract
- Protein synthesis is a major energy-consuming process of the cell that requires the controlled production1-3 and turnover4,5 of ribosomes. Although the past few years have seen major advances in our understanding of ribosome biogenesis, structural insight into the degradation of ribosomes has been lacking. Here we present native structures of two distinct small ribosomal 30S subunit degradation intermediates associated with the 3' to 5' exonuclease ribonuclease R (RNase R). The structures reveal that RNase R binds at first to the 30S platform to facilitate the degradation of the functionally important anti-Shine-Dalgarno sequence and the decoding-site helix 44. RNase R then encounters a roadblock when it reaches the neck region of the 30S subunit, and this is overcome by a major structural rearrangement of the 30S head, involving the loss of ribosomal proteins. RNase R parallels this movement and relocates to the decoding site by using its N-terminal helix-turn-helix domain as an anchor. In vitro degradation assays suggest that head rearrangement poses a major kinetic barrier for RNase R, but also indicate that the enzyme alone is sufficient for complete degradation of 30S subunits. Collectively, our results provide a mechanistic basis for the degradation of 30S mediated by RNase R, and reveal that RNase R targets orphaned 30S subunits using a dynamic mechanism involving an anchored switching of binding sites.