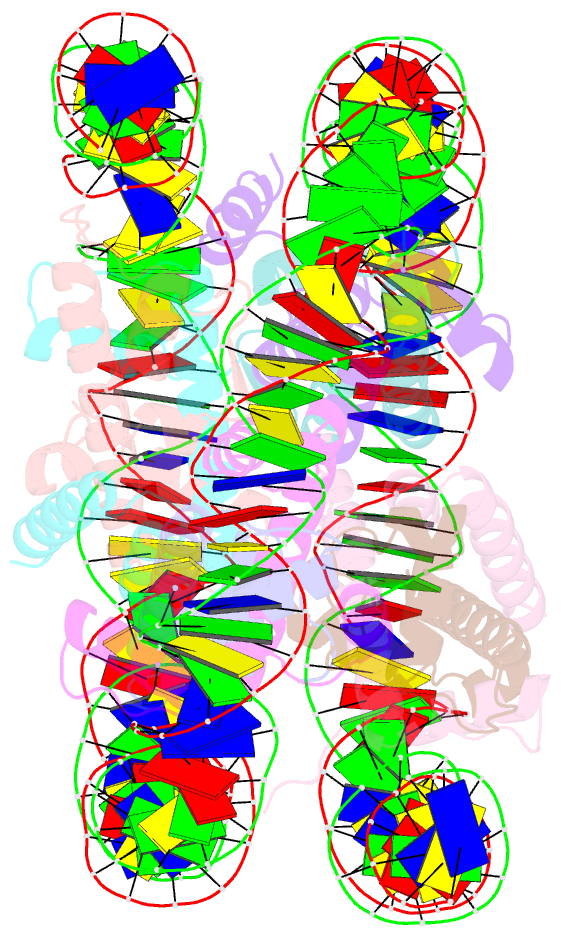
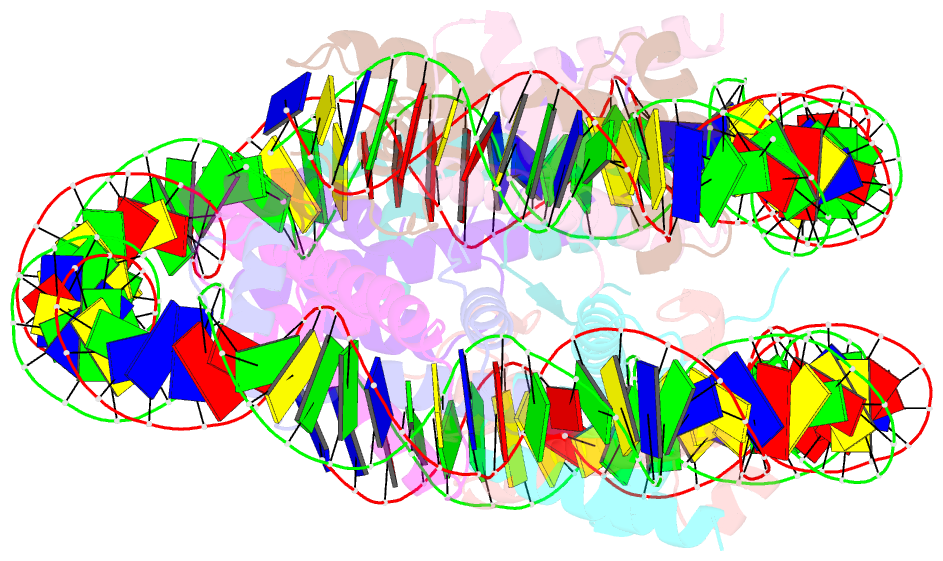
Summary information and primary citation
- PDB-id
- 8com; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- DNA binding protein
- Method
- cryo-EM (3.3 Å)
- Summary
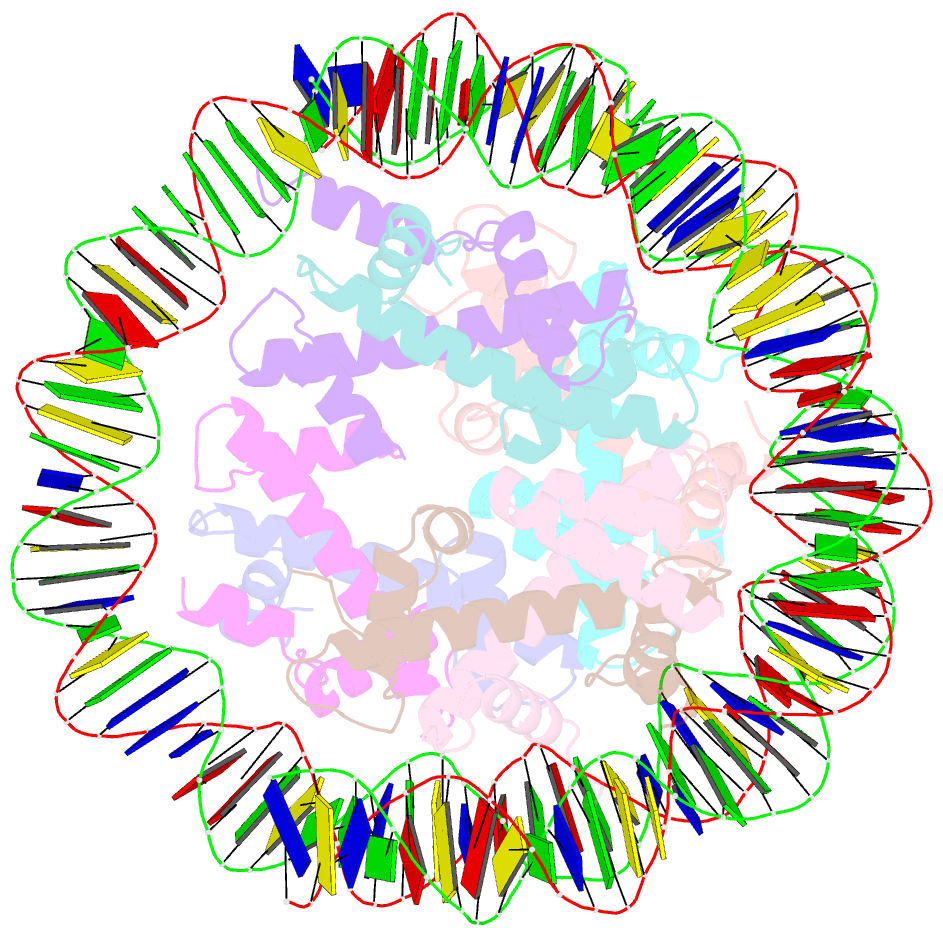
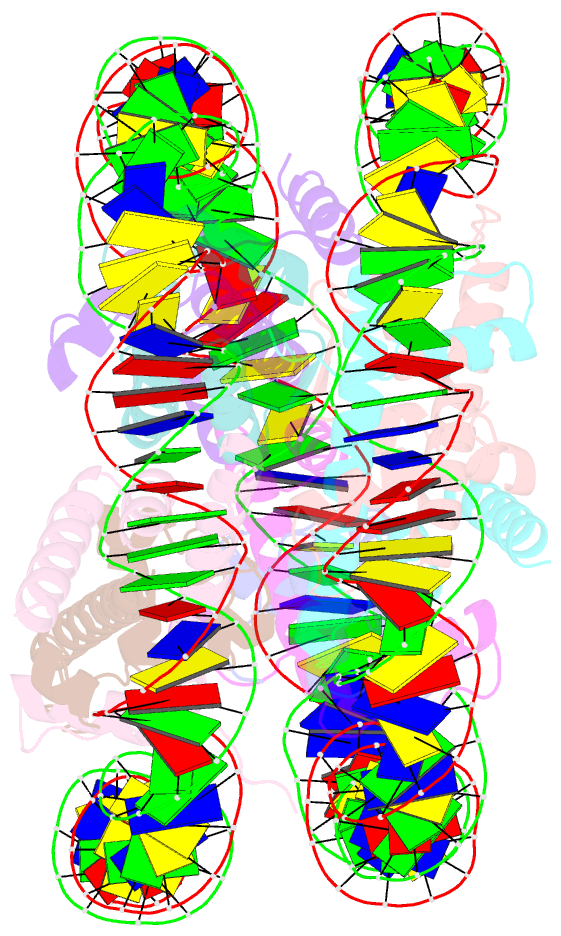
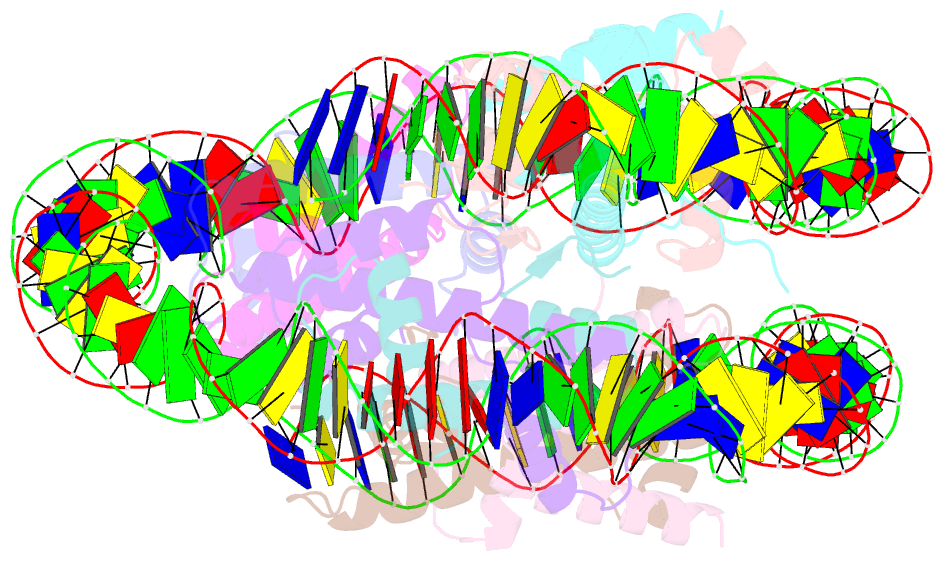
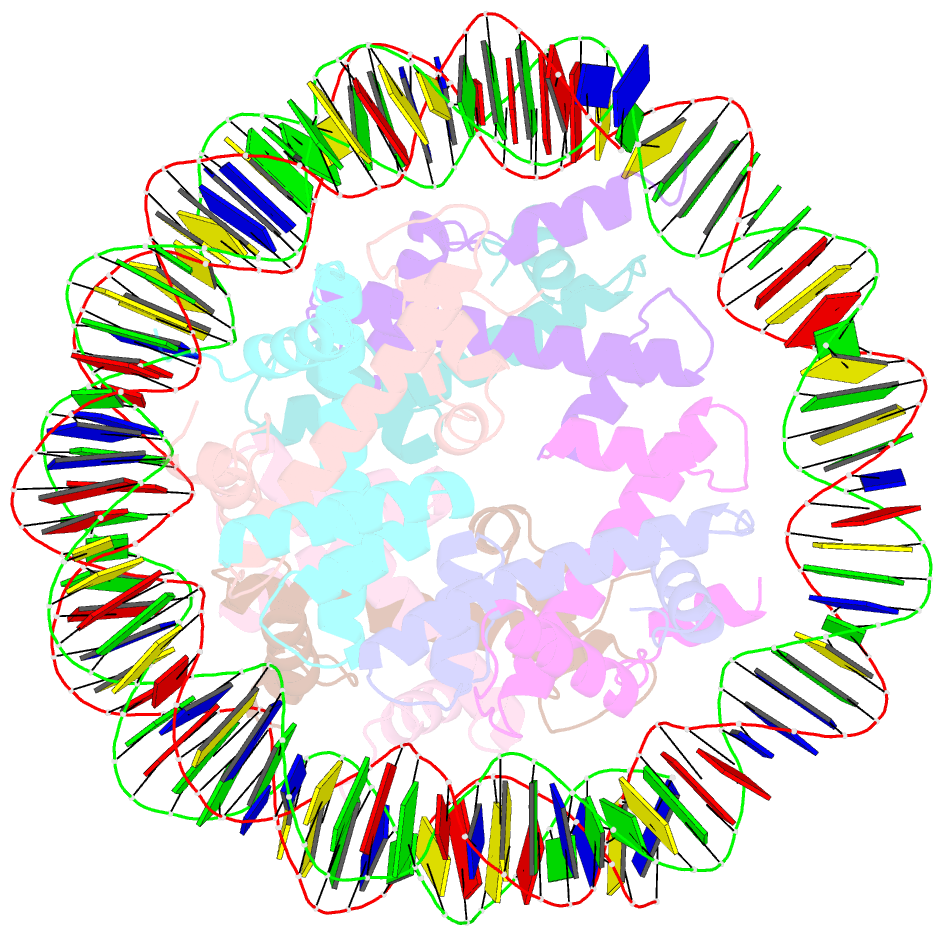
- Structure of the nucleosome core particle from trypanosoma brucei
- Reference
- Deak G, Wapenaar H, Sandoval G, Chen R, Taylor MRD, Burdett H, Watson JA, Tuijtel MW, Webb S, Wilson MD (2023): "Histone divergence in trypanosomes results in unique alterations to nucleosome structure." Nucleic Acids Res., 51, 7882-7899. doi: 10.1093/nar/gkad577.
- Abstract
- Eukaryotes have a multitude of diverse mechanisms for organising and using their genomes, but the histones that make up chromatin are highly conserved. Unusually, histones from kinetoplastids are highly divergent. The structural and functional consequences of this variation are unknown. Here, we have biochemically and structurally characterised nucleosome core particles (NCPs) from the kinetoplastid parasite Trypanosoma brucei. A structure of the T. brucei NCP reveals that global histone architecture is conserved, but specific sequence alterations lead to distinct DNA and protein interaction interfaces. The T. brucei NCP is unstable and has weakened overall DNA binding. However, dramatic changes at the H2A-H2B interface introduce local reinforcement of DNA contacts. The T. brucei acidic patch has altered topology and is refractory to known binders, indicating that the nature of chromatin interactions in T. brucei may be unique. Overall, our results provide a detailed molecular basis for understanding evolutionary divergence in chromatin structure.